Fluorescent quantitative PCR detection method for drug resistance gene mcr (mobile colistin resistance)-4/5/8
A technology of drug resistance genes and primer pairs, which can be used in microorganism-based methods, biochemical equipment and methods, and determination/inspection of microorganisms, which can solve problems such as time-consuming and labor-intensive
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Embodiment 1, design and preparation of primers
[0062] A large number of sequence analyzes and comparisons were carried out to obtain several primers for detecting the drug resistance gene mcr-4 / 5 / 8. Preliminary experiments were carried out on each primer to compare performances such as sensitivity and specificity, and finally a primer set for detecting the drug resistance gene mcr-4 / 5 / 8 was obtained, as shown in Table 1.
[0063] Table 1 Primer information table of mcr-4 / 5 / 8 gene
[0064]
[0065] Primer F1 and primer R1 form primer pair I;
[0066] Primer F2 and primer R2 constitute primer pair II;
[0067] Primer F3 and primer R3 constitute primer pair III.
Embodiment 2
[0068] Embodiment 2, the construction of plasmid standard
[0069] 1. mcr-4 plasmid standard product: insert the double-stranded DNA molecule shown in sequence 7 of the sequence table into the EcoR V site of pDM19-T vector to obtain the mcr-4 plasmid standard product.
[0070] 2. mcr-5 plasmid standard product: insert the double-stranded DNA molecule shown in Sequence 8 of the sequence table into the EcoR V site of pDM19-T vector to obtain the mcr-5 plasmid standard product.
[0071] 3. mcr-8 plasmid standard product: insert the double-stranded DNA molecule shown in sequence 9 in the sequence table into the EcoRV site of pDM19-T vector to obtain the mcr-8 plasmid standard product.
Embodiment 3
[0072] Embodiment 3, primer verification and specificity experiment
[0073] 1. Plasmid Standard Verification Primers
[0074] Samples to be tested: 3 plasmid standards prepared in Example 2.
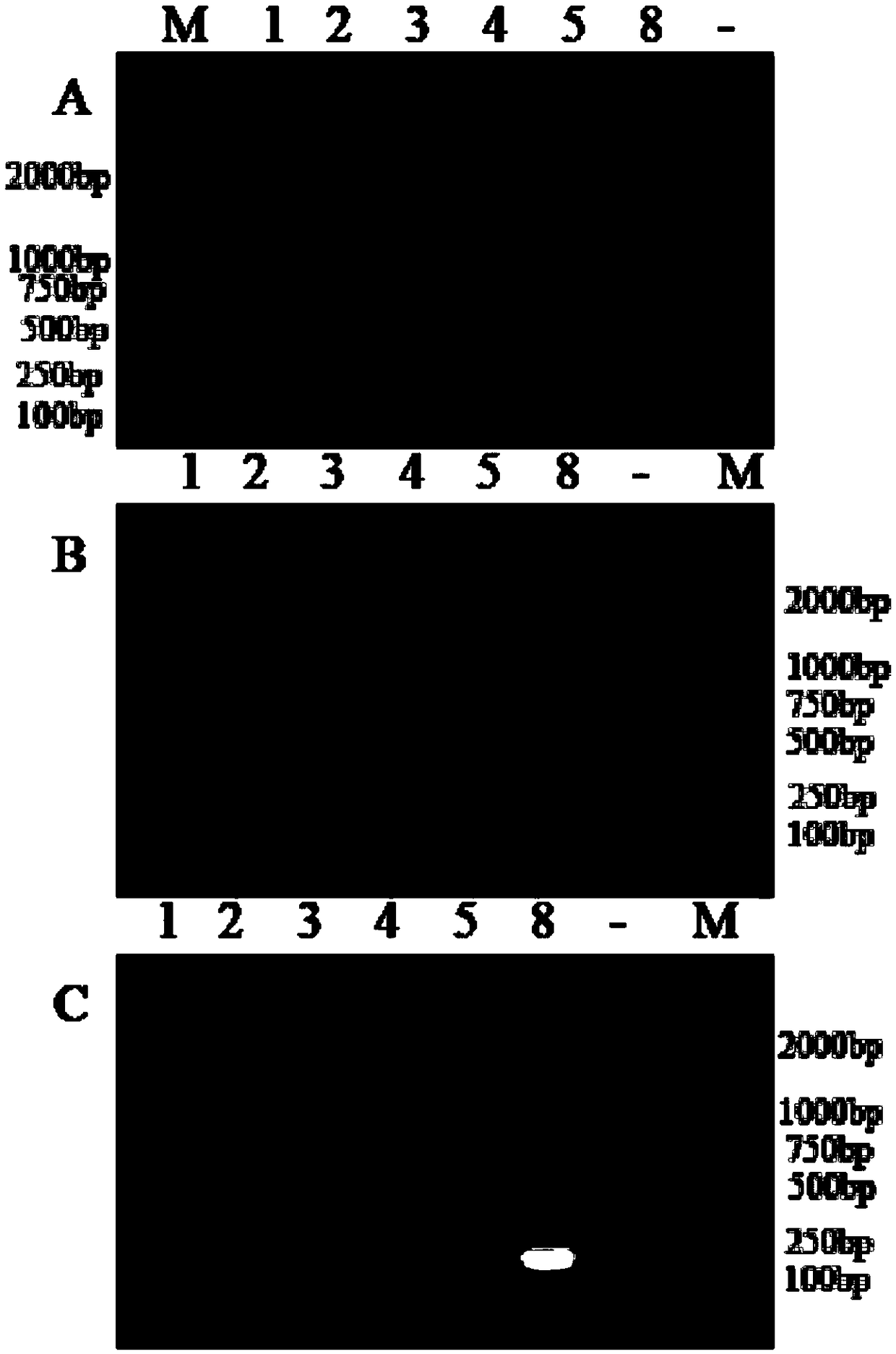
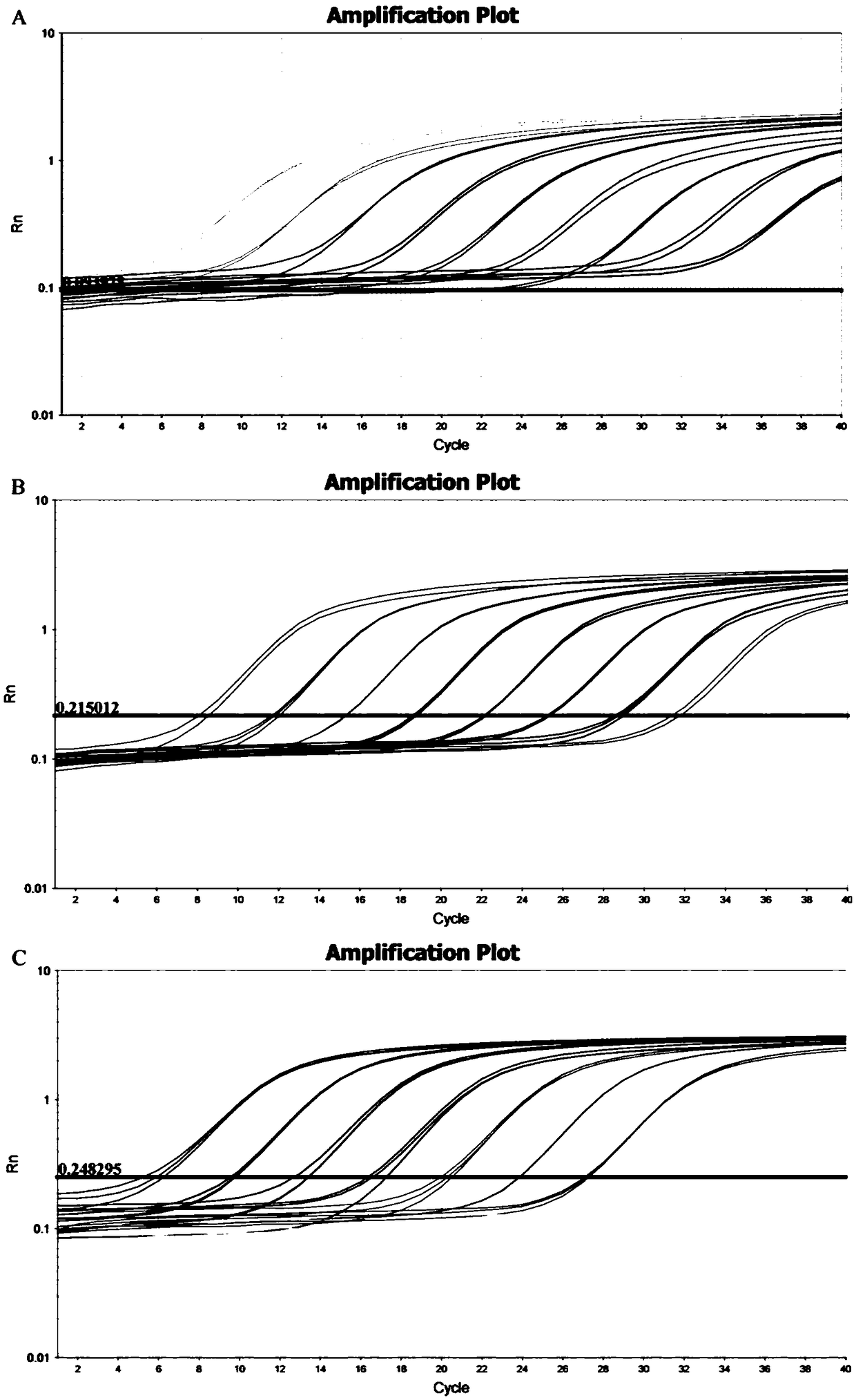
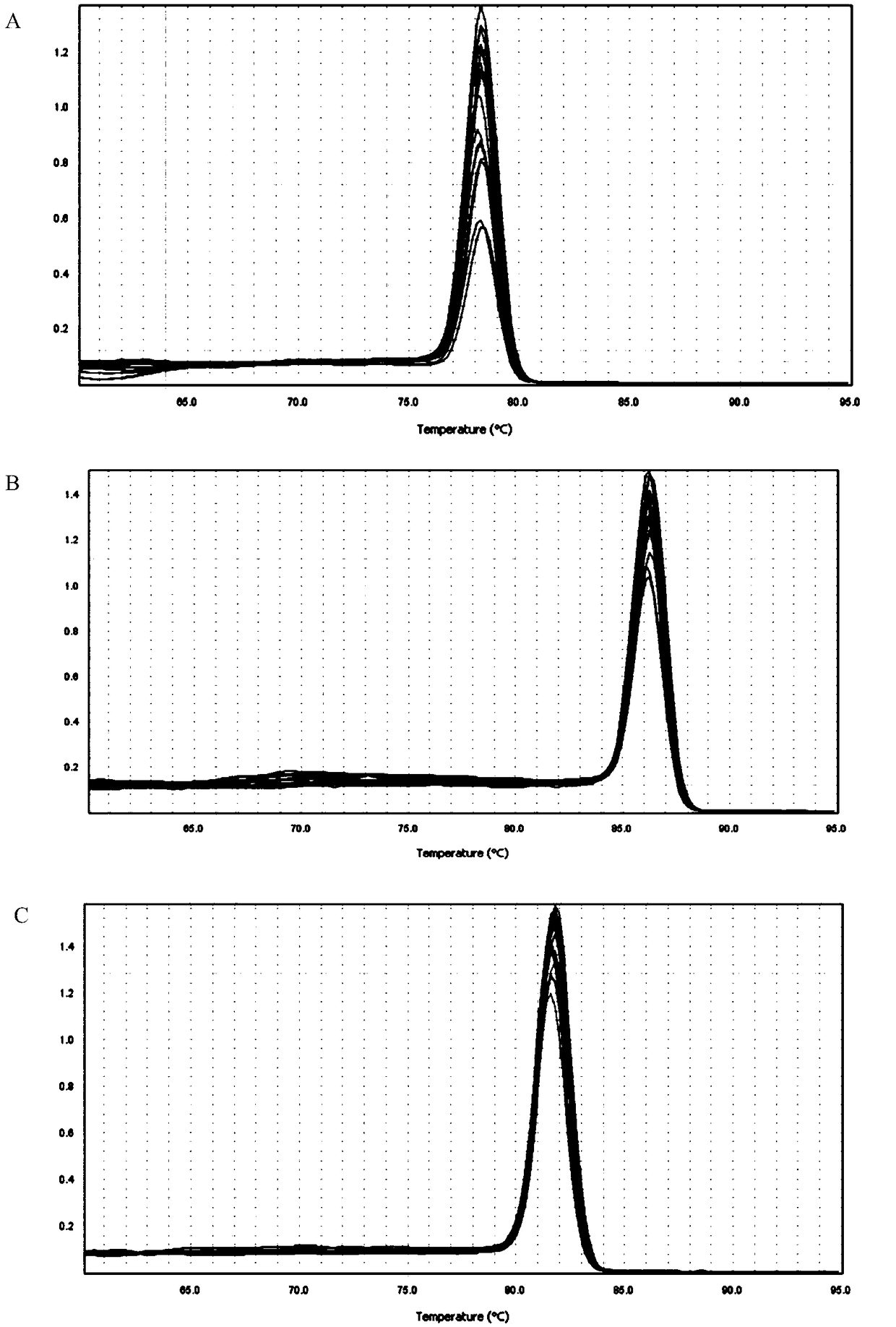
[0075] Using the sample to be tested as a template, PCR amplification was performed using primer pair I to primer pair III in Example 1, respectively.
[0076] PCR amplification reaction system (20 μl): upstream primer 0.8 μl, downstream primer 0.8 μl, 2×Taq PCRMasterMix (purchased from Nanjing Novizan Biotechnology Co., Ltd., P111-01) 10 μl, nucleic acid-free water 6.4 μl, DNA template 2 μl .
[0077] The concentrations of upstream primers and downstream primers in the system are both 0.4 pmol / μL.
[0078] Nucleic acid-free water was used as a negative control.
[0079] PCR amplification reaction program: pre-denaturation at 95°C, 5min; 30× (denaturation at 95°C, 30s; annealing at 60°C, 30s; extension at 72°C, 30s); extension at 72°C, 10min.
[0080] The results showed that the pr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com