Method for changing feeding habits of bombyx mori and application of method
A technology of silkworm and feeding habits, applied in genome editing and biological fields
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Embodiment 1, the cloning of hestia gene
[0050] The genome sequence of the target gene hestia is as follows (SEQ ID NO: 9, wherein the underlined sequence is an exon sequence, and the non-underlined sequence is an intron sequence):
[0051] ATGTCACCACCGCTAGTCCATATCAATACATTTGTTCAACCCAAGCAAAGTACACTGTAGACAAAAGTA TCAAAGTTTTTTATAATATGTAGTTTTTCTTAGGCGTTAATAGGTTGCCTATTATATCATCGAAACATGTGTATAC AATTCCTTCTATAATTTATACTTTCGTACTAATGTGTGTACTAAATTTTTTCGGGTTTGATTCTGTCTCATTATCTA TCATGAGTTTGAACCTTGTATTACATATCTTATGTTCCTTCCTTGGTATGTTTTTTTTGGAAGAGAATGCGCCTGTAT TATTCAGAGCTCTGTAAGTTTGATATTTGCATCGGATGCAGACCAATAACTGCACAAGGTTCCAGTAAACTTGTAAT TCAGACTTGCATTATAAATGTTTTGATAGCTTTGGTGTTTATTGTGCCGAATTCACTTCAGATTCTAATCAAACCAG TTATATATTTGCTTCCCATGCACGCCTTTGTGTCGTTCGAAGTGCATTATTATGGCCACCTTCTCAATTTACTTATC CCGCGTTTACATTTAATAAACTATTACATGGAATCCTCATTAACTACCACAAGCGATAAAAGGGAGTCGAGTGTACT GAAACATGTTATTTTTATTTAAATATTATAATAAGGAATCGAACTGTCAAATGAAAAAATTTATGGATCTTTATTATA TTATCGTAGAATTCTTACAGATATCTTA...
Embodiment 2
[0059] Embodiment 2, the selection of sgRNA target (TS) and the synthesis of sgRNA
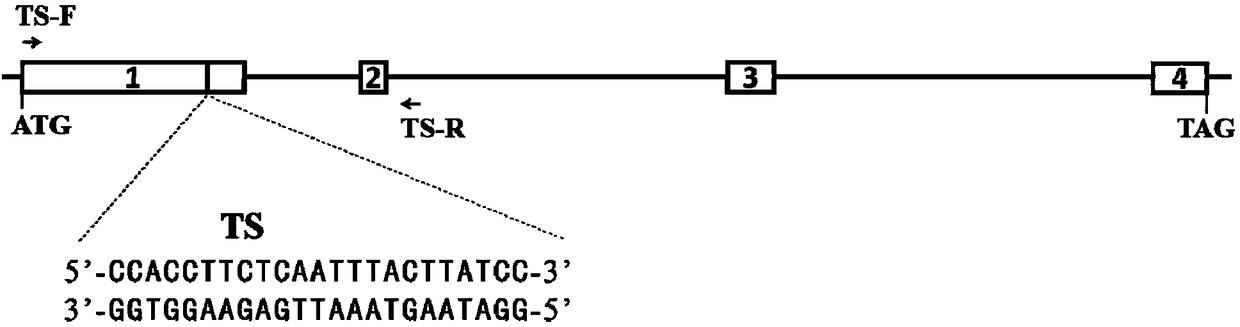
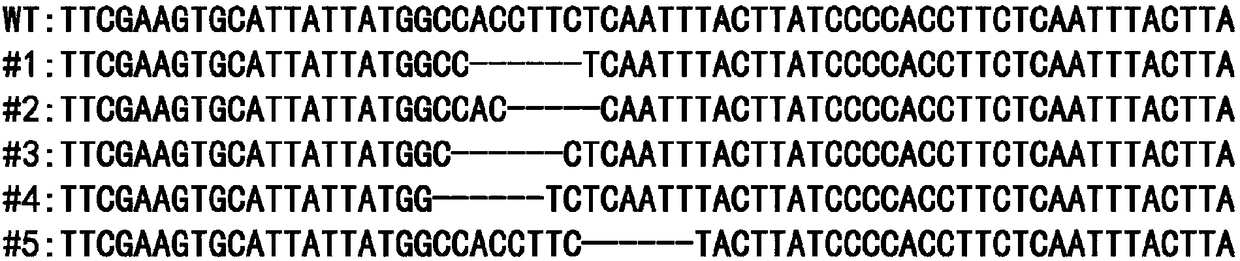
[0060] The present invention uses the CRISPR / Cas9 system to knock out the hestia gene, and screens out a 23bp sgRNA recognition target (TS) located in exon 1 after extensive selection and testing of the gene sequence. Schematic diagram of the structure of the silkworm hestia gene and its target (TS) sequence as shown in figure 1 shown.
[0061] The sequence of the sgRNA recognition target is as follows:
[0062] 5'-GGATAAGTAAATTGAGAAGGTGG-3' (SEQ ID NO: 1).
[0063] sgRNA framework sequence (SEQ ID NO:11):
[0064] GNNNNNNNNNNNNNNNNNNNNN GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTT
[0065] Wherein, the underlined part is the sgRNA targeting sequence.
[0066] According to the screened target (TS), the following primers sgF and sgR were synthesized:
[0067] sgF:
[0068] TAATACGACTCACTATAGGATAAGTAAATTGAGAAGGGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAG ...
Embodiment 3
[0073] Embodiment 3, the acquisition of Cas9 mRNA
[0074] The template plasmid used to synthesize Cas9 mRNA is pTD1-Cas9 (Yueqiang Wang, Zhiqian Li, JunXu, Baosheng Zeng, lin ling, Lang You, Yazhou Chen, Yongping Huang*, and Anjiang Tan* .The CRISPR / Cas System mediates efficient genome engineering in Bombyxmori. Cell Research 23(12):1414-6.2013). The nucleotide sequence of the plasmid is shown as SEQ ID NO: 8, the first ATG is a start codon, and the last TAG is a stop codon. A nuclear localization signal precedes the stop codon.
[0075] The pTD1 vector contains the T7 promoter, 5'-UTR and 3'-UTR sequences, and the ORF of the Cas9 gene is connected between the 5'-UTR and 3'-UTR sequences, so pTD1-Cas9 can be used as a template for in vitro transcription of Cas9 mRNA . The pTD1-Cas9 vector was linearized with Not I restriction endonuclease, and the gel was collected and purified as a template for Cas9 mRNA synthesis. Use mMESSAGEmMACHINE T7 kit to transcribe mRNA in vit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com