An automated method for detecting reassortment of segmented RNA viruses
A technology of segments and viruses, applied in the field of automatic detection of segmented RNA virus reassortment, can solve problems such as wrong reassortment detection results, unreasonable genome evolution relationship, etc. The method is simple and effective, the detection speed is fast, and unreasonable avoidance Effects of Sex and Complexity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042]Example 1, virus reassurance detection method
[0043]I. Preparation of the specific classification of each segment after the release
[0044]For the same variety of virus strains, the method of detecting the virus is included in the following steps:
[0045]1. Build a viral genome segment database
[0046]The genomic nucleic acid sequence of all intact genomics and the genomic nucleic acid sectors of the same virus strain are obtained from the database.
[0047]The sequence of each segment of the genomic nucleic acid sequence of different virus strains is used to perform multi-sequence alignment to obtain a multi-segment alignment result of each of the versions of all virus strains, which is recorded as a viral genome segment database.
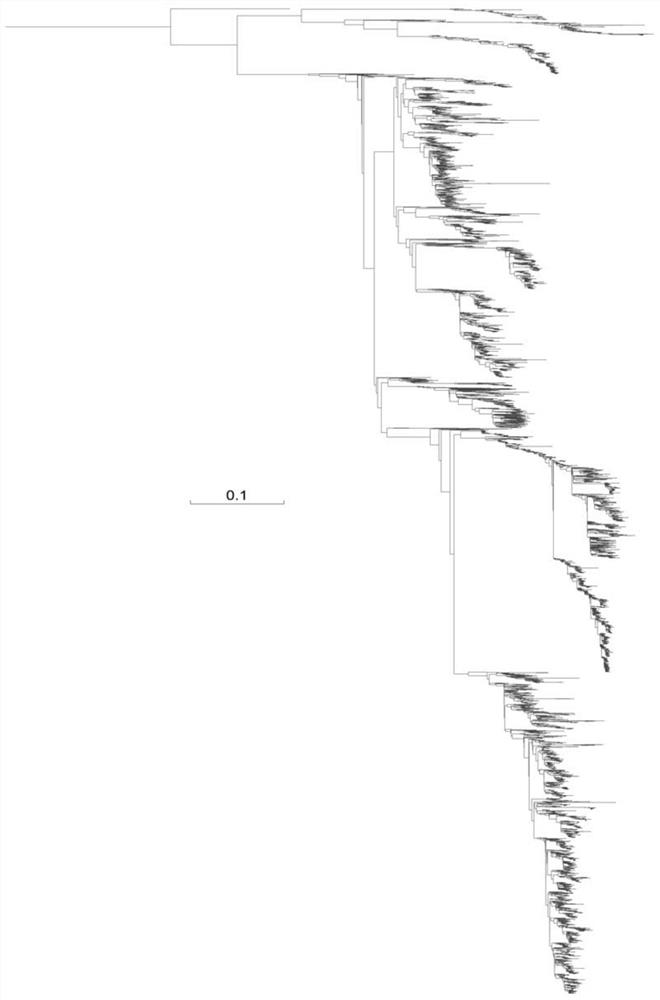
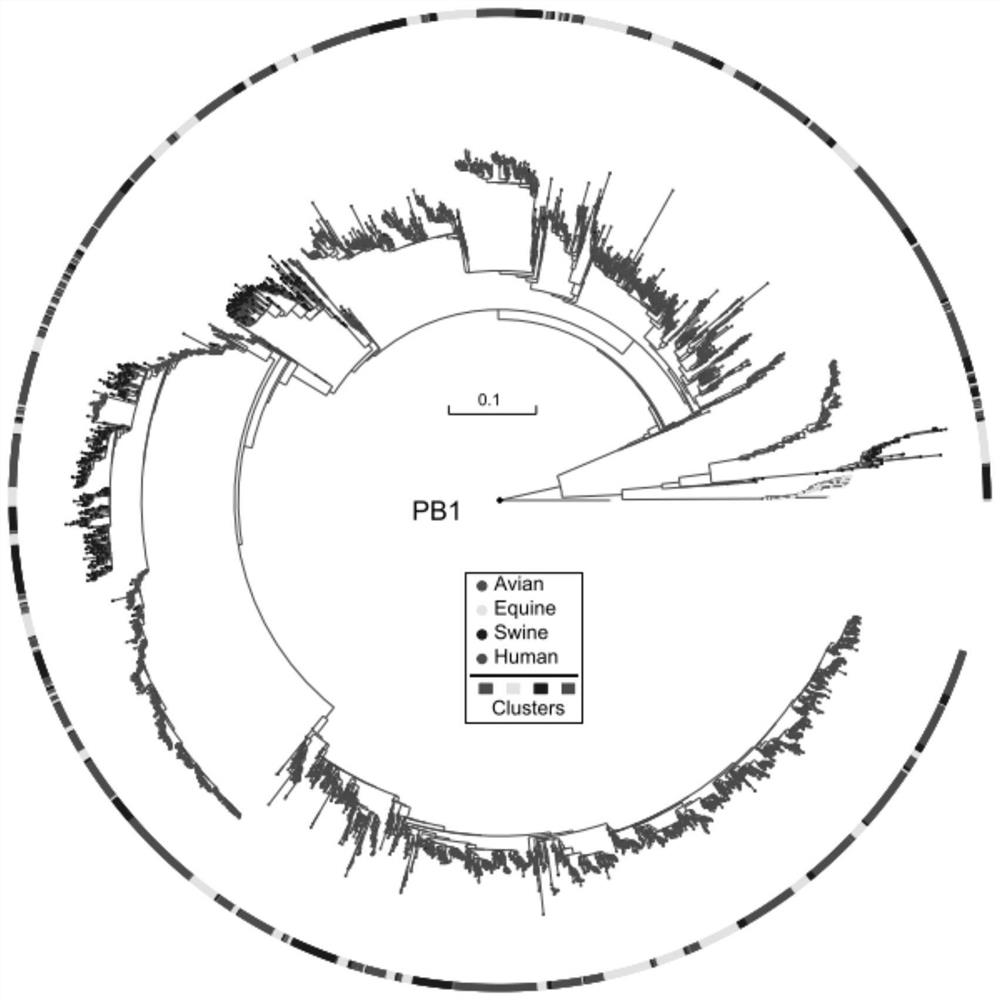
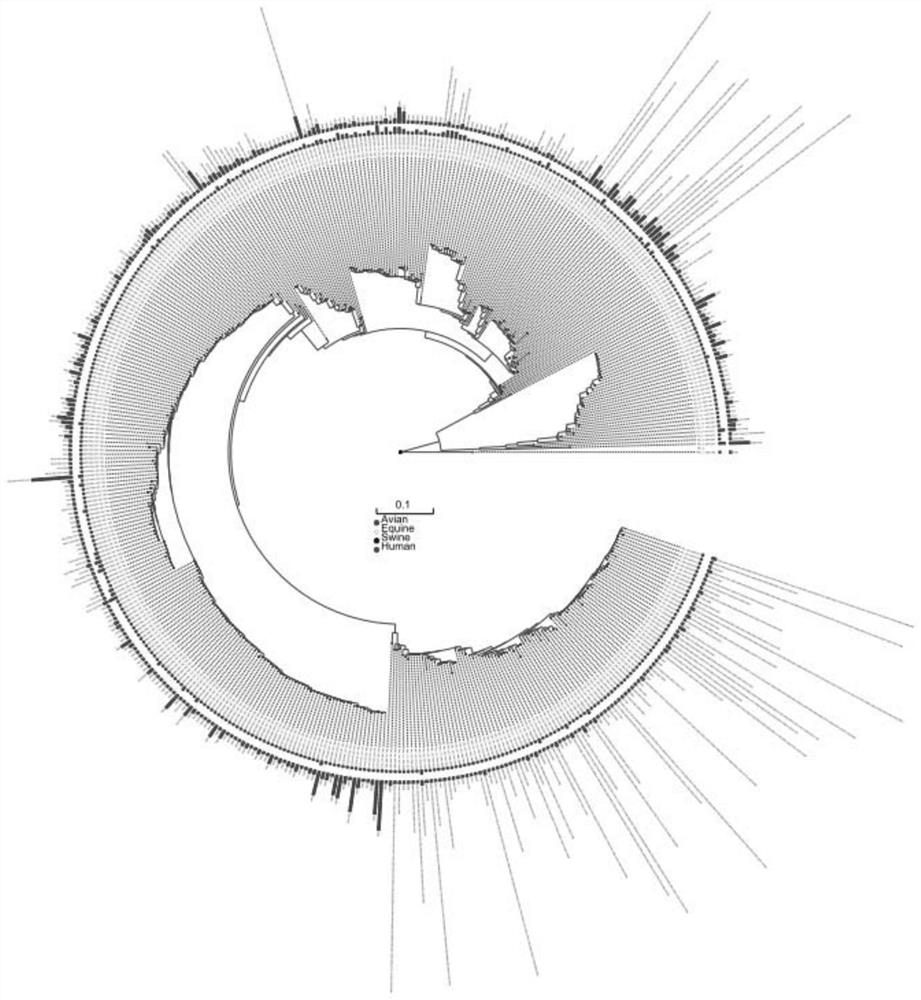
[0048]2, build system development trees
[0049]The multi-segment comparison result of each of the versions of all virus strains obtained in step 1 is constructed by system development trees by maximum likelihood method, and the system development tree of each se...
Embodiment 2
[0077]Example 2, influenza virus reassurance detection
[0078]1, download the gisaid website to download the full influenza virus genome sequence data
[0079]Log in to gisaid (http: / / platform.gisaid.org), filtering the downloading of the hypertrophic virus genome nucleic acid sequence containing a complete genome and full length sequence, generates download files, downloading each influenza virus genome sequence data; There is also a genome such as a genome such as illegal characters, and delete the redundant genome.
[0080]In this embodiment, the full influenza genome sequence data downloaded in the Gisaid website database in 2016-10-26, with a total of 18,564 viral genome after data quality control.
[0081]The sequence of each segment of the genomic algorithm sequence of different virus strains utilizes the MAFFT software for multi-sequence alignment to obtain a multi-segment alignment result of each of the versions of all virus strains, which is recorded as a viral genome segment databas...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com