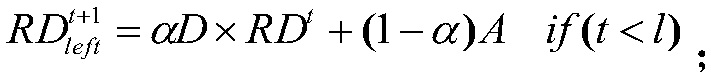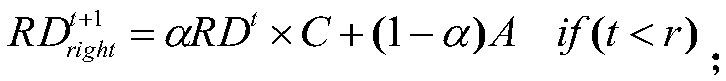Drug susceptibility prediction method based on multi-similarity network
A drug sensitivity and prediction method technology, applied in the field of biomedicine, can solve the problems of inaccurate prediction of drug sensitivity and failure to consider the characteristics of biological networks, so as to achieve accurate prediction and improve the accuracy rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
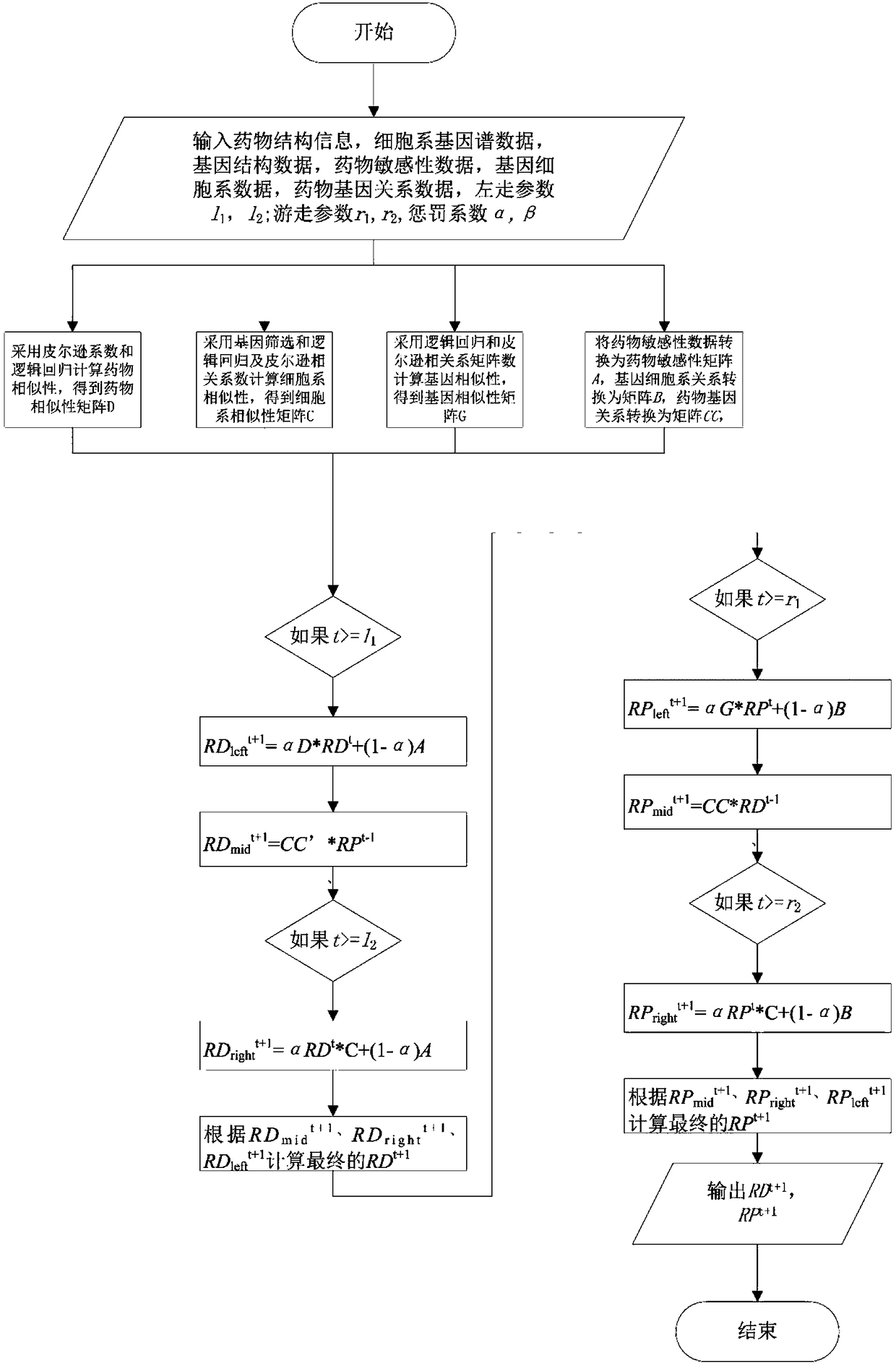
[0027] The present invention first constructs three similarity networks: drug similarity network (DSN), cell line similarity network (CSN) and gene similarity network (GSN), and the mapping relationship among them. The construction of DSN is to find the 1D&2D structure file SDF file of the compound corresponding to the drug in the database, and then use PaDEL software to analyze the descriptor of the drug, and finally calculate the Pearson correlation coefficient of the descriptor of these compounds to obtain the drug similarity network. The construction is to form a cell line similarity network by calculating the Pearson correlation coefficient of the gene spectrum data corresponding to the cell line in the data set. As for the gene similarity network (GSN), we first obtain the gene names obtained from the drug-target (gene) relationship, and then obtain the gene's interaction network from the HPRD database. Drug sensitivity is then predicted based on the three biological net...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com