A primer set and dual RT-PCR method for the detection of bovine norovirus and bovine coronavirus
A coronavirus and primer set technology, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc., to achieve the effect of improving detection efficiency, accurate detection results, and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1: Construction of a double RT-PCR method for the detection of bovine norovirus and bovine coronavirus
[0035] 1. Design and synthesis of primer sets for the detection of bovine norovirus and bovine coronavirus
[0036]The sequences of bovine norovirus and bovine coronavirus published in GenBank were compared and analyzed, and the ORF1 gene of bovine norovirus (the accession number in Genbank was NC_029645) and the bovine coronavirus (the accession number in Genbank were NC_029645) were selected respectively. A segment of the ORF1ab gene of U00735) is the target sequence amplified by each virus, and Primer 5.0 is used to design primers for the above target sequence. The amplification primers of the two designed viruses were analyzed by DNAstar to avoid the formation of stable primer-dimers; at the same time, the homology or complementarity of the amplified sequences of each pair of primers was analyzed. Analysis to avoid high homology or complementarity betwee...
Embodiment 2
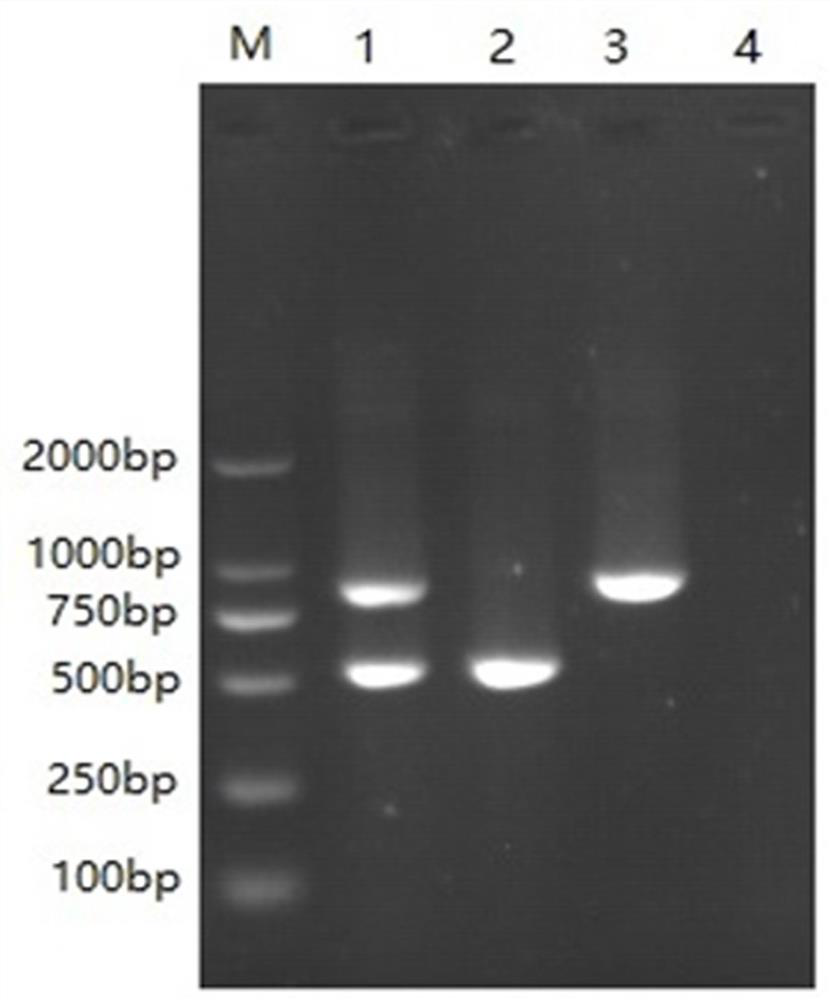
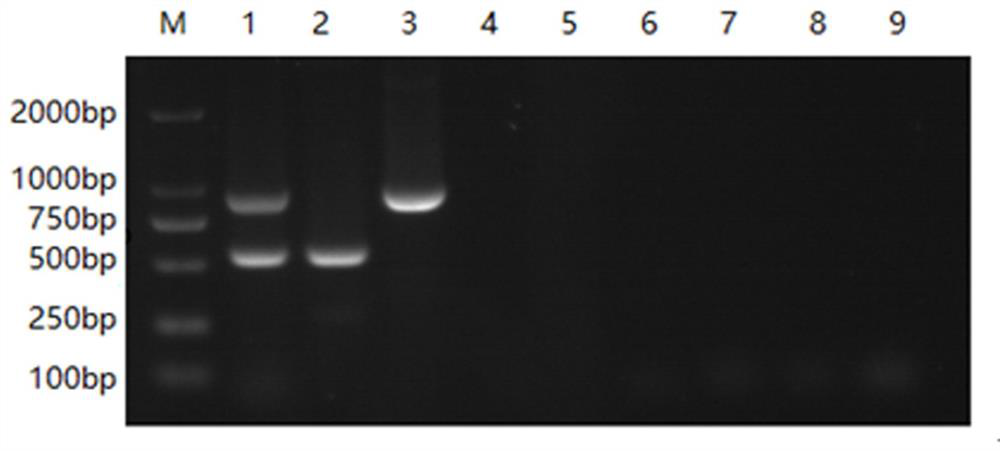
[0052] Example 2: Specificity Experiment
[0053] Utilize the double RT-PCR method for detecting bovine norovirus and bovine coronavirus constructed in Example 1 of the present invention to detect bovine norovirus, bovine coronavirus, BRV (bovine rotavirus), BAstV (bovine astrovirus), BVDV ( Bovine viral diarrhea virus), BKV (bovine crista virus), BToV (bovine ring torovirus) were detected, and a negative control (with ddH 2 (2), verify the specificity of the double RT-PCR method established by the present invention, and the concrete method is as follows:
[0054] BRV (Bovine Rotavirus), BAstV (Bovine Astrovirus), BVDV (Bovine Viral Diarrhea Virus), BKV (Bovine Crest Virus), BToV (Bovine Cyclovirus) were extracted according to the method described in Step 2 in Example 1. virus) RNA, respectively reverse transcription to obtain each virus cDNA;
[0055] Take the mixed cDNA of bovine norovirus and bovine coronavirus, the cDNA of bovine norovirus, the cDNA of bovine coronavirus...
Embodiment 3
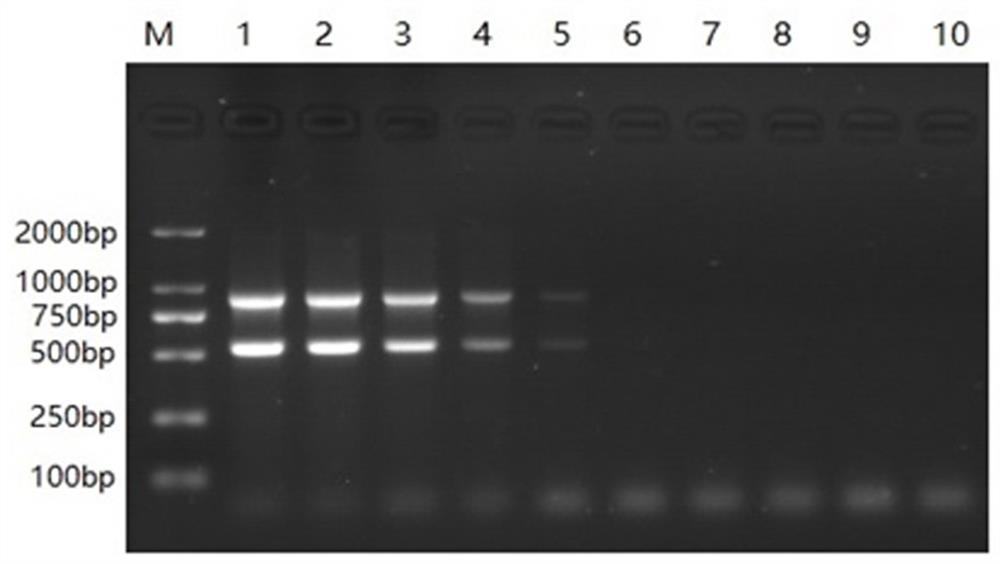
[0056] Example 3: Sensitivity experiment
[0057] The DNA concentration of bovine norovirus was 23.8 ng / μL, and the DNA concentration of bovine coronavirus was 54.45 ng / μL, according to the formula (6.02×10 23 copy number / mol)×(nucleic acid concentration ng / μL×10 -9 ) / (nucleic acid length × 660) = standard plasmid copy number copies / μL, the number of copies of bovine norovirus DNA is calculated to be 6.71 × 10 9 copies / μL, the copy number of bovine coronavirus DNA is 1.38×10 10 copies / μL; 0.5 μL of bovine coronavirus DNA and 0.5 μL of bovine norovirus DNA were added to 4 μL of sterile double-distilled water at the same time, and mixed to obtain a mixture of bovine coronavirus DNA and bovine norovirus DNA (bovine coronavirus DNA and bovine norovirus DNA). In the mixture of coronavirus DNA and bovine norovirus DNA, the copy number of bovine norovirus DNA is 6.71×10 8 copies / μL, the copy number of bovine coronavirus DNA is 1.38×10 9 copies / μL); then the mixture of bovine noro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com