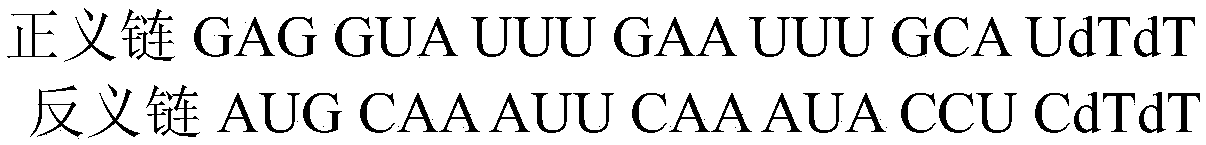Method for analyzing load rate of graphene oxide for nucleic acid
An analysis method, graphene technology, applied to the analysis of materials, material excitation analysis, material analysis through optical means, etc., to achieve the effect of simple calculation and convenient operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1: An example of analyzing the loading rate of GO to a kind of green fluorescent double-stranded RNA.
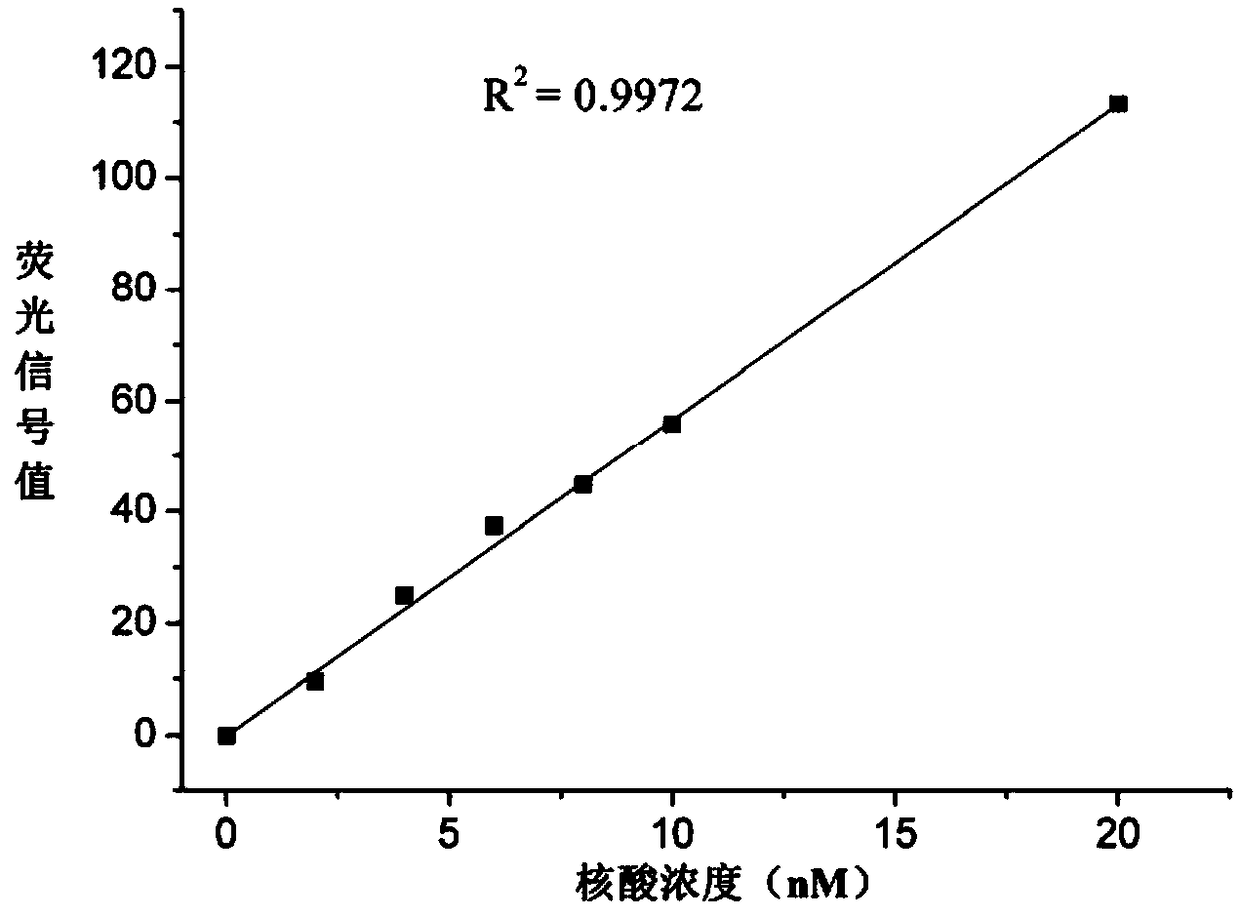
[0025] The sequence of the double-stranded RNA is shown in the appendix figure 2 As shown, the molar mass M is 12112 g / mol, the fluorescent modification group is FAM, the emission wavelength is 480 nm, and the excitation wavelength is 520 nm.
[0026] The specific implementation process is as follows: prepare DEPC solutions with concentrations of 0, 2, 4, 6, 8, 10, and 20 nM of the nucleic acid respectively, and use a fluorescence spectrophotometer (Shanghai Prism Technology Co., Ltd., model F96PRO, emission wavelength 480 nm, Excitation wavelength 520nm) for detection, obtain the standard fluorescence signal values of the nucleic acid solution to be tested at each concentration, and draw a standard curve ( figure 1 ). Bring the volume to 1.6 mL with an initial concentration of n 1 The DEPC solution of the nucleic acid (treated with high-temperature and...
Embodiment 2
[0030] Example 2: An example of analyzing the loading rate of GO on a red fluorescent double-stranded RNA.
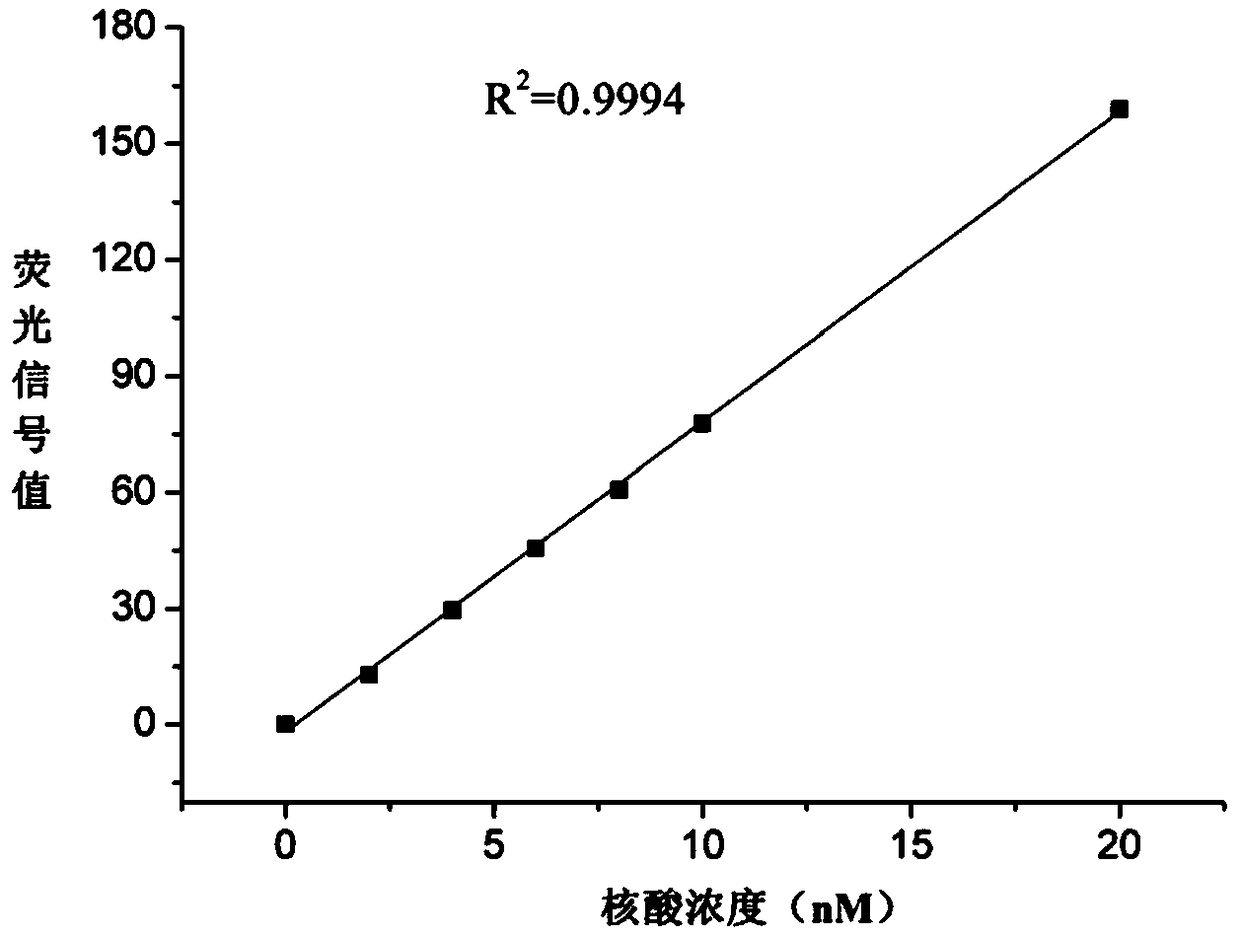
[0031] The sequence of the double-stranded RNA is attached Figure 4 , the molar mass M is 12112 g / mol, the fluorescent modification group is CY3, the emission wavelength is 550 nm, and the excitation wavelength is 570 nm.
[0032] The specific implementation process is as follows: prepare DEPC solutions with concentrations of 0, 2, 4, 6, 8, 10, and 20 nM of the nucleic acid respectively, and use a fluorescence spectrophotometer (Shanghai Prism Technology Co., Ltd., model F96PRO, emission wavelength 550 nm, Excitation wavelength 570nm) for detection, obtain the standard fluorescence signal values of the nucleic acid solution to be tested at each concentration, and draw a standard curve ( figure 2 ). Bring the volume to 1.6 mL with an initial concentration of n 1 The DEPC solution of the nucleic acid and the DEPC solution of GO (20 μg / mL) were mixed in equal volume...
Embodiment 3
[0036] Example 3: An example of analyzing the loading rate of GO to a kind of green fluorescent single-stranded DNA.
[0037] The single-stranded DNA sequence is attached Figure 6 , the molar mass M is 23016 g / mol, the fluorescent modification group is FAM (green fluorescence), the emission wavelength is 480 nm, and the excitation wavelength is 520 nm.
[0038] The specific implementation process is as follows: prepare TE solutions with concentrations of 0, 2, 4, 6, 8, 10, and 20 nM of the nucleic acid respectively, and use a fluorescence spectrophotometer (Shanghai Prism Technology Co., Ltd., model F96PRO, emission wavelength 480 nm, Excitation wavelength 520 nm) for detection, obtain the standard fluorescence signal values of the nucleic acid solution to be tested at each concentration, and draw a standard curve ( image 3 ). Bring the volume to 1.6 mL with an initial concentration of n 1 The TE solution of the nucleic acid and the TE solution of GO (20 μg / mL) were mix...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com