miRNA-340 target gene binding sequence, recombinant plasmid and application thereof
A technology that combines sequences and recombinant plasmids, applied in DNA/RNA fragments, genetic engineering, recombinant DNA technology, etc., can solve problems such as intractable diseases and adverse reactions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Cloning of Example 1 Mouse IL-17A 3'UTR Nucleic Acid Sequence
[0057] Step S101: The nucleic acid sequence of mmu-IL-17A 3'UTR found in NCBI is from the 535th base to the 1171st base in the IL-17A gene (as shown in SEQ ID No.1), and the designed primers are:
[0058] F: 5'-CGGCTAGCCGACAGAGACCCGCGGCTGACCCCTAAG-3' (as shown in SEQ ID No.2);
[0059] R: 5'-CCGCTCGAGCGGTTTAAAACAGAGTAGGGAGCT-3' (shown in SEQ ID No. 3).
[0060] Step S102: Using mouse genomic DNA as a template, using the nucleotide sequences shown in SEQ ID No.2 and SEQ ID No.3 as primers, using high-fidelity enzymes to perform conventional PCR reaction amplification, and gel after electrophoresis The PCR product was recovered, and NheI and XhoI were used for double digestion, and the digested product was recovered and purified.
Embodiment 2
[0061] Example 2 Construction of the recombinant reporter plasmid of pmir-GLO-IL-17A 3'UTR complete fragment
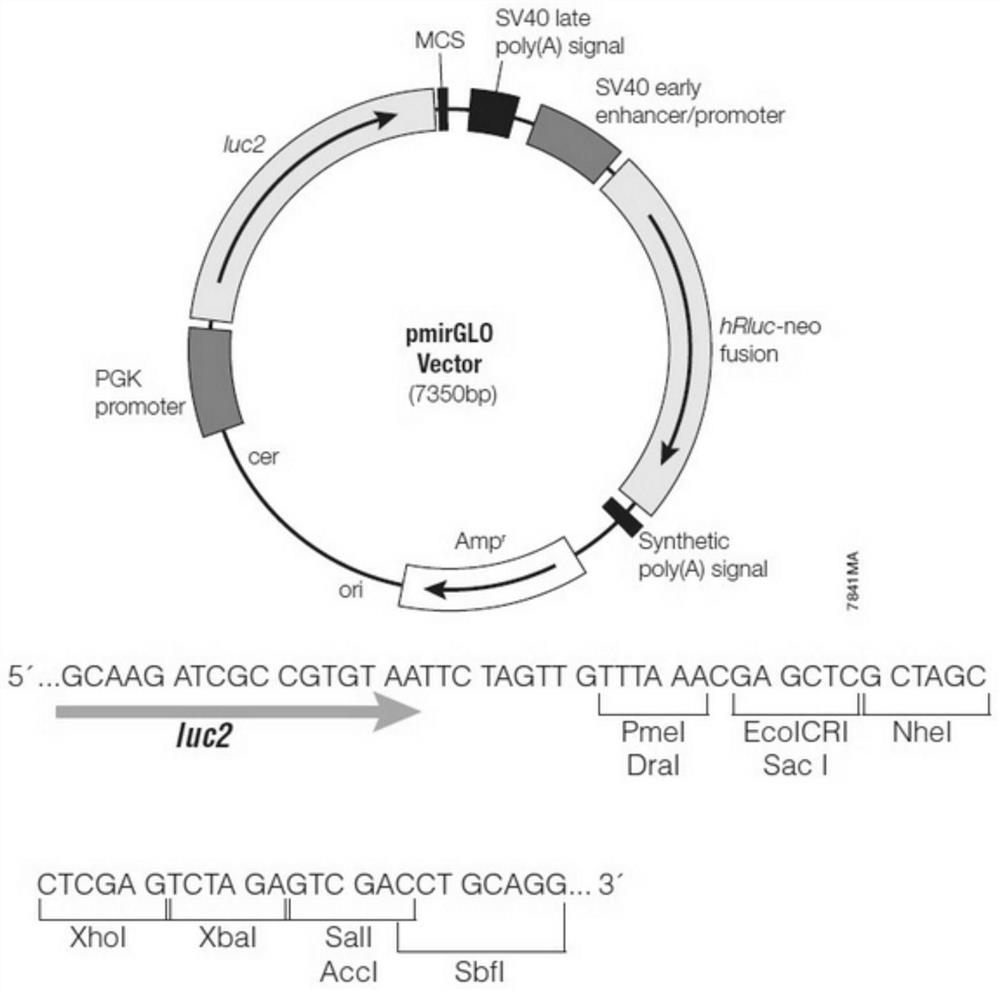
[0062] Step S201: Transform conventionally prepared DH5α competent cells with the pmir-GLO plasmid for amplification, prepare the pmir-GLO plasmid with a mini-prep kit, check the purity and quality of the plasmid by electrophoresis, perform double digestion of the plasmid with NheI and XhoI, and gel The recovery kit recovers the digested products of large fragments; the pmir-GLO plasmid map is as follows figure 1 As shown, the pmir-GLO plasmid in this embodiment is the pmirGLO Dual-Luciferase miRNA Target Expression vector (abbreviated as pmirGLO).
[0063] Ligation, transformation and clonal amplification of recombinant plasmids
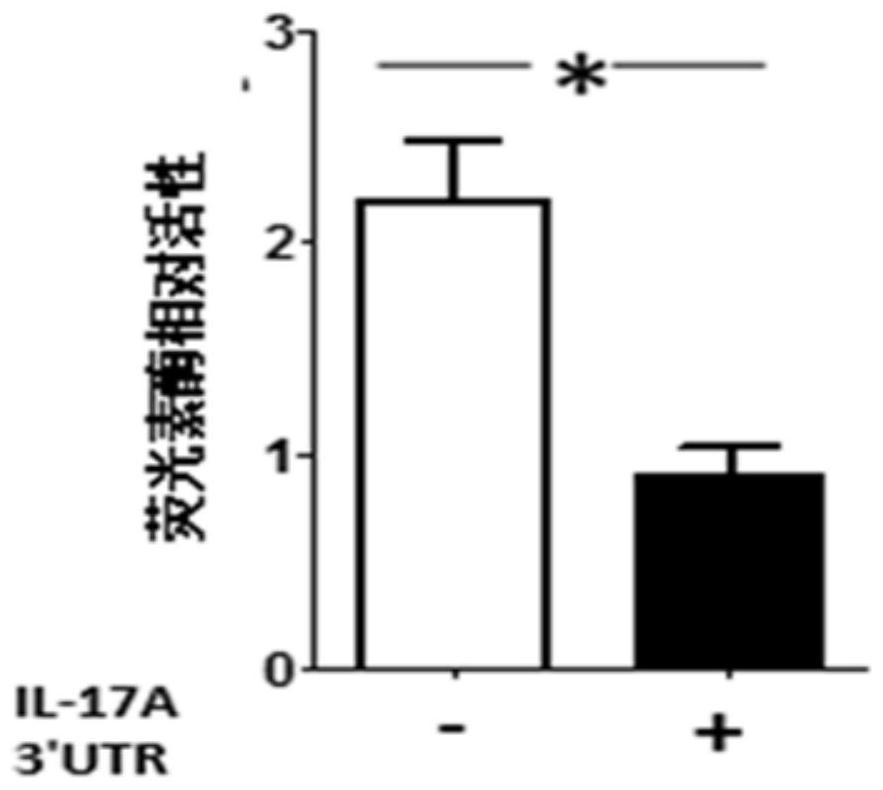
[0064] Step S202: Perform a ligation reaction between the digested mmu-IL-17A 3'UTR PCR product prepared in Example 1 and the pmir-GLO plasmid.
[0065] Step S203: The ligation product is transformed into competent cells DH5α, selectively...
Embodiment 3I
[0067] Example 3 Construction of IL-17A 3'UTR fragment deletion reporter plasmid
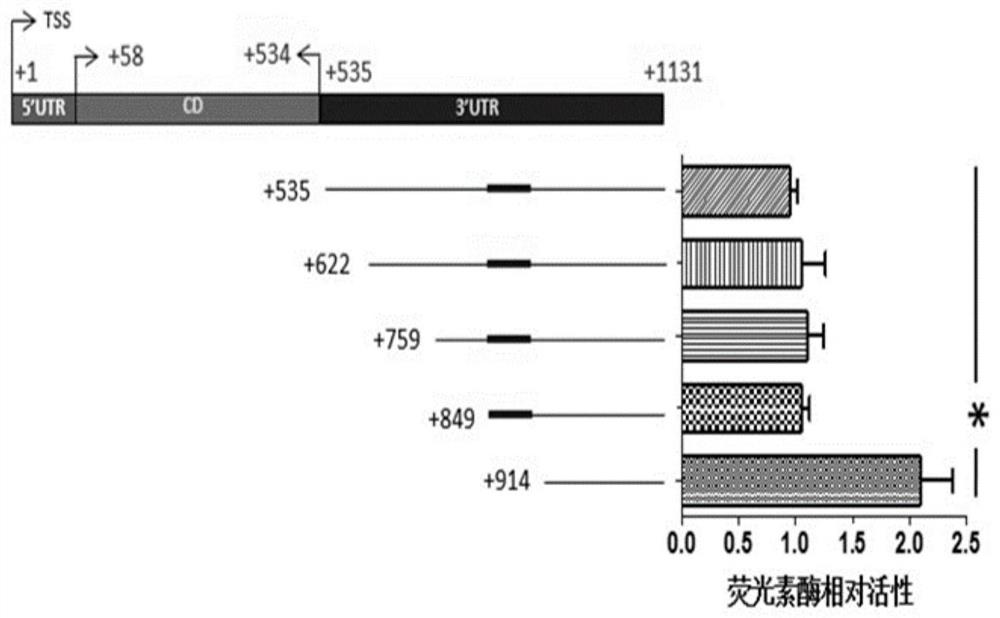
[0068] S301: Design the different primers shown in Table 1, and perform PCR using the constructed pmir-GLO-IL-17A 3'UTR complete fragment recombination reporter plasmid in Example 2 as a template to obtain a series of IL-17A deletions of different lengths 3'UTR fragments: F1(+535~+1131), F2(+622~+1131), F3(+759~+1131), F4(+849~+1131), F5(+914~+1131), Wherein, F1 is the 535th base to the 1131st base in the IL-17A gene (as shown in SEQ ID No.4), and F2 is the 622nd to the 1131st base in the IL-17A gene (as shown in SEQ ID No. .5), F3 is the 759th base to the 1131st base in the IL-17A gene (as shown in SEQ ID No.6), and F4 is the 849th to the 1131st base in the IL-17A gene ( As shown in SEQ ID No.7), F5 is the 914th base to the 1131st base in the IL-17A gene (as shown in SEQ ID No.8).
[0069] Table 1 Primers for IL-17A 3'UTR deletion fragment
[0070]
[0071] Step S302: agarose gel electrop...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com