Growth-related molecular marker in C-type scavenger receptor of Litopenaeusvannamei and application
A technology related to molecular markers and scavenger receptors, applied in the determination/testing of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the problem of low marker density, no genes or markers directly related to growth and other issues to achieve the effect of improving efficiency and accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0017] Example 1: Obtaining growth-related molecular markers of Litopenaeus vannamei
[0018] (1) Genome-wide association analysis (GWAS) of growth traits of Litopenaeus vannamei:
[0019] The material used for GWAS analysis was a mixed population consisting of 13 full-sib families. In July 2015, a total of 13 full-sib families were constructed in Hainan Guangtai Ocean Breeding Co., Ltd. (Wenchang, Hainan) through directional mating. After each family hatched the larvae, they were transferred to 5m 2 The ponds are cultivated separately. During this period, in order to reduce the impact of the breeding environment on different families, we tried our best to adopt a standardized way to cultivate each family, so that the conditions such as salinity, temperature, larvae density, bait and aeration were as consistent as possible. In September 2015, 50 prawns were randomly selected from each of 13 families and mixed and transferred to 10m 2 The pools were cultivated in a unified ...
Embodiment 2
[0036] Example 2: Application of a growth-related marker of Litopenaeus vannamei in breeding.
[0037] (1) Typing of the LvSRC-211-G / T loci of the candidate parents
[0038] In June 2016, using the reserved broodstock of Hainan Guangtai Ocean Breeding Company (Wenchang, Hainan) as the candidate parents, the DNA was extracted from a pair of swimming legs of each prawn, and the following primers were used to amplify the DNA of each individual Target area sequence:
[0039] LvSRCF: CCGTATTTGTTGAATGAGTGGG
[0040] LvSRCR: CTACCACTCCAACAGACGAAGG
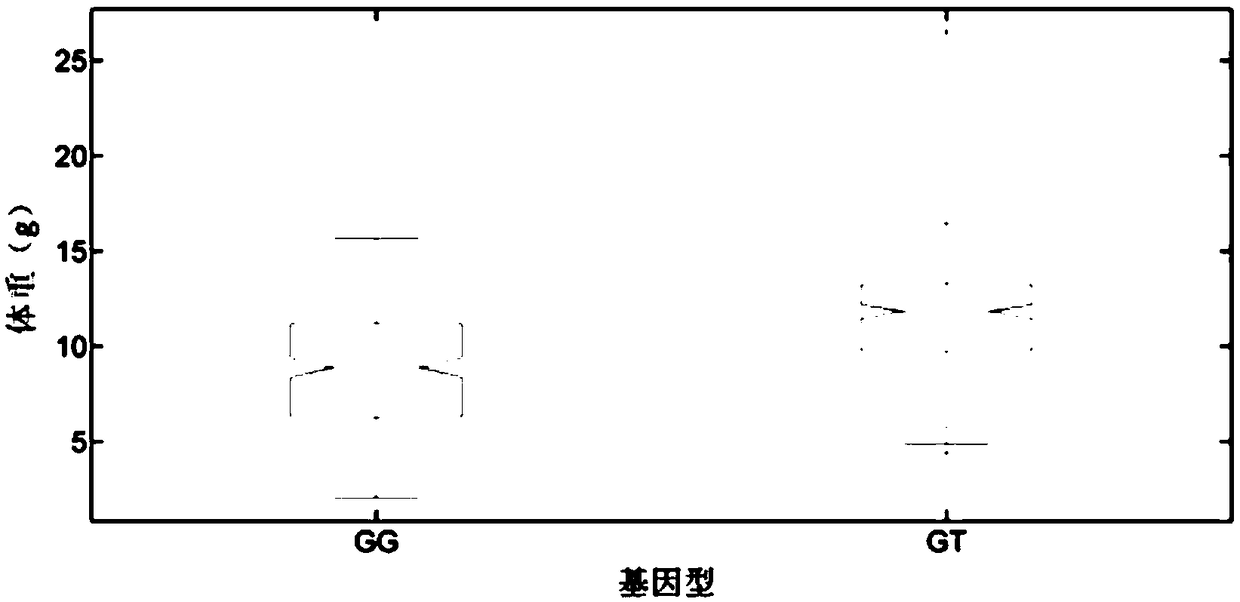
[0041] The PCR amplification products were sequenced using ABI3730xl sequencer, and the sequencing primers were LvSRCF primers, and the LvSRC-211-G / T was typed according to the sequencing peak map. If there was only one G base peak at this position, the typing result It is GG type. If there is only one T base peak, the typing result is TT type. If there are both G base peaks and T base peaks at this position, the typing result is GT ty...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com