DNA polymerase with increased gene mutation specificity and PCR buffer composition for increasing activity thereof
A gene mutation and polymerase technology, applied in the field of DNA polymerase, can solve problems such as inability to obtain clear information and poor synthesis of polymerase, and achieve improved amplification efficiency, high matching extension selectivity, and reliable gene mutation-specific amplification Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0133] Example 1 Inducing Mutation of Taq Polymerase
[0134] 1-1. Fragment PCR
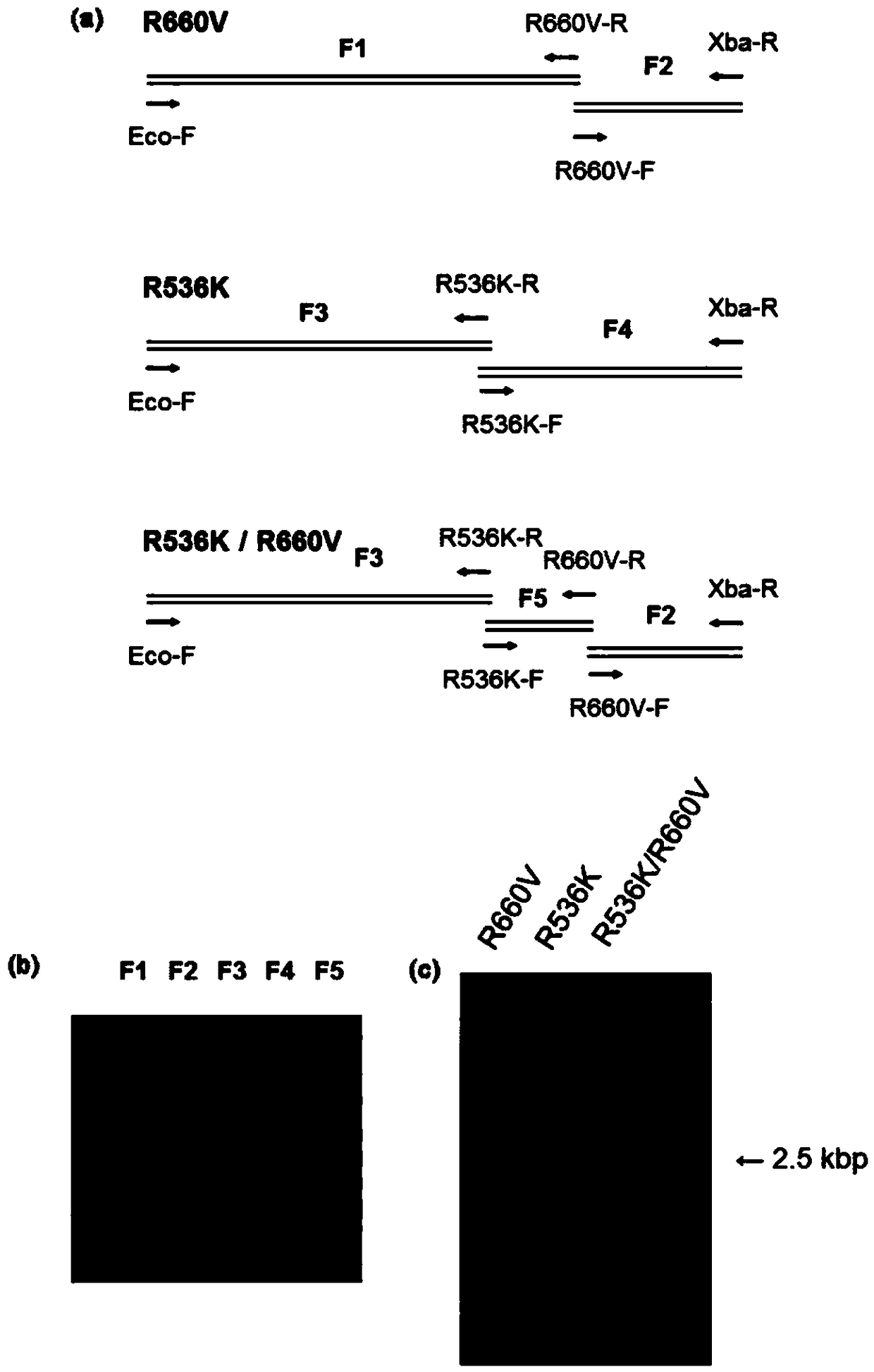
[0135] In this example, in the amino acid sequence with SEQ ID NO: 1, the arginine at the 536th amino acid residue (in the amino acid sequence of SEQ ID NO: 1 was prepared according to the following method) was substituted by lysine for Taq DNA polymerization enzyme (hereinafter referred to as "R536K"), Taq DNA polymerase in which the arginine at the 660th amino acid residue was replaced with valine (hereinafter referred to as "R660V"), and the arginine at the 536th amino acid residue was replaced with Taq DNA polymerase in which lysine was substituted and arginine at the 660th amino acid residue was substituted by valine (hereinafter referred to as "R536K / R660V") was prepared as follows.
[0136] First, Taq DNA polymerase fragments (F1 to F5) were amplified by PCR using the mutation-specific primers shown in Table 1, as figure 1 (a) shown. The reaction conditions are shown in Table 2.
[013...
Embodiment 2
[0163] Example 2 Introduction of E507K mutation
[0164] 2-1. Fragment PCR
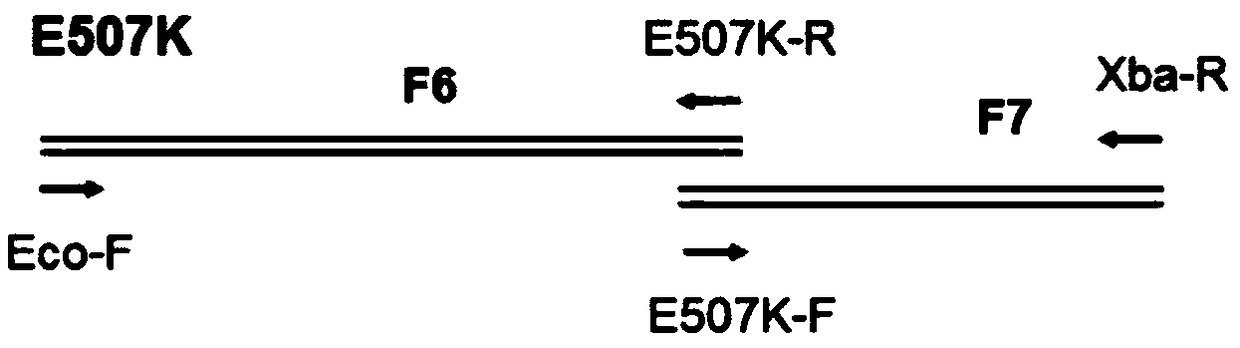
[0165] The activity of the Taq polymerase of "R536K", "R660V" and "R536K / R660V" prepared in Example 1 was tested, and the results confirmed that the activity decreased (data not shown), respectively for "R536K", "R660V " and "R536K / R660V" introduced the E507K mutation (in the amino acid sequence of SEQ ID NO: 1, the glutamic acid at the 507th amino acid residue was replaced by lysine), as a control group, in the wild-type (WT) Taq DNA polymer The E507K mutation was also introduced into the enzyme. The preparation method of the TaqDNA polymerase with the E507K mutation introduced is the same as in Example 1.
[0166] Use the mutation-specific primers shown in Table 9, such as image 3 Taq DNA polymerase fragments (F6 to F7) were amplified by PCR as indicated. The reaction conditions are shown in Table 10.
[0167] Table 9
[0168]
[0169] Table 10
[0170]
[0171] *Template plasmid: pUC1...
Embodiment 3
[0184] Example 3 Using the DNA polymerase of the present invention to carry out qPCR
[0185] Using Taq polymerases each containing the "E507K / R536K", "E507K / R660V" and "E507K / R536K / R660V" mutations obtained in Example 2 above, it was confirmed that it extends the mismatched primer for the template containing the SNP. whether the ability is reduced. As a control, "E507K" Taq polymerase containing the E507K mutation was used.
[0186] The templates containing SNPs used in this example are rs1408799, rs1015362 and rs4911414. The genotypes of each template and the sequence information of their corresponding specific primers (IDT, USA) are shown in Table 12 and Table 13.
[0187] Table 12
[0188]
[0189] Table 13
[0190]
[0191] The qPCR conditions (Applied Biosystems 7500 Fast) are shown in Table 14 below.
[0192] Table 14
[0193]
[0194] The probes were double labeled as shown in Table 15 below.
[0195] Table 15
[0196]
[0197] Oral epithelial cells ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com