Establishing method of blue honeysuckle sequence-related amplified polymorphism (SRAP) reaction system
A reaction system and blue fruit technology, applied in the field of molecular biology, can solve problems such as reduction of amplification products, reduction of polymorphic bands, reduction of primer-template binding rate, etc., and achieve the effect of simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
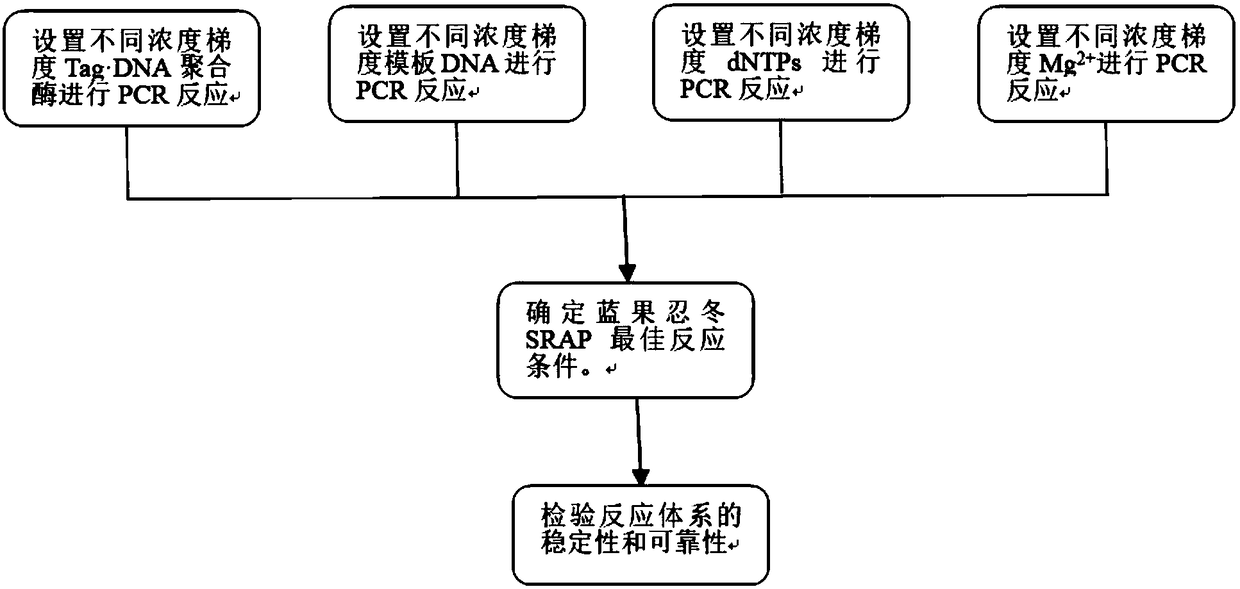
[0028] Example 1: (1) The system was optimized by a single factor test. The basic reaction conditions were a total reaction volume of 20 μl, of which 10×PCR buffer 2 μl, MgCl2 2.5 mmol / l, dNTPs 0.15 mmol / l, primer 0.1 μmol / l, template DNA 20ng, Tag DNA polymerase 1.5U, make up the insufficient part with double distilled water, and carry out the reaction.
[0029] (2) On the basis of the basic reaction conditions in step (1), set the concentration of Tag DNA polymerase to 0.5U for reaction. Take 10 μl of the final product of the reaction and add 1 μl of 6× loading buffer to mix, and electrophoresis in 1.8% agarose gel. The electrophoresis buffer was 1×TAE, the electrophoresis electric field strength was 5V / cm, stained with Gofdview, observed, photographed and recorded under the Alphalmager HP gel imaging system. Select the concentration of Tag DNA polymerase with clear and bright bands.
[0030](3) On the basis of the basic reaction conditions in step (1), set the template DN...
example 2
[0035] Example 2: (1) The system was optimized by a single factor test. The basic reaction conditions were a total reaction volume of 20 μl, of which 10×PCR buffer 2 μl, MgCl2 2.5 mmol / l, dNTPs 0.15 mmol / l, primer 0.1 μmol / l, template DNA 20ng, Tag DNA polymerase 1.5U, make up the insufficient part with double distilled water, and carry out the reaction.
[0036] (2) On the basis of the basic reaction conditions in step (1), set the concentration of Tag DNA polymerase to 1.0 U for reaction. Take 10 μl of the final product of the reaction and add 1 μl of 6× loading buffer to mix, and electrophoresis in 1.8% agarose gel. The electrophoresis buffer was 1×TAE, the electrophoresis electric field strength was 5V / cm, stained with Gofdview, observed, photographed and recorded under the Alphalmager HP gel imaging system. Select the concentration of Tag DNA polymerase with clear and bright bands.
[0037] (3) On the basis of the basic reaction conditions in step (1), set the template ...
example 3
[0042] (1) The system was optimized by a single factor test. The basic reaction conditions were a total reaction volume of 20 μl, including 2 μl of 10×PCR buffer, 2.5 mmol / l of MgCl2, 0.15 mmol / l of dNTPs, 0.1 μmol / l of primers, and 20 ng of template DNA , Tag DNA polymerase 1.5U, make up the insufficient part with double distilled water, and carry out the reaction.
[0043] (2) On the basis of the basic reaction conditions in step (1), set the concentration of Tag DNA polymerase to 1.5 U for reaction. Take 10 μl of the final product of the reaction and add 1 μl of 6× loading buffer to mix, and electrophoresis in 1.8% agarose gel. The electrophoresis buffer was 1×TAE, the electrophoresis electric field strength was 5V / cm, stained with Gofdview, observed, photographed and recorded under the Alphalmager HP gel imaging system. Select the concentration of Tag DNA polymerase with clear and bright bands.
[0044] (3) On the basis of the basic reaction conditions in step (1), set t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com