Paired sgRNA for gene editing and application thereof
A gene editing and gene technology, applied in the field of gene editing, can solve problems such as obstacles, and achieve the effect of technical flexibility, high efficiency, and good homogeneity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Example 1 Improved paired Cas9-sgRNA gene editing method
[0049] 1. The establishment of a theoretical system based on the precise NHEJ-based paired Cas9-sgRNA gene editing method
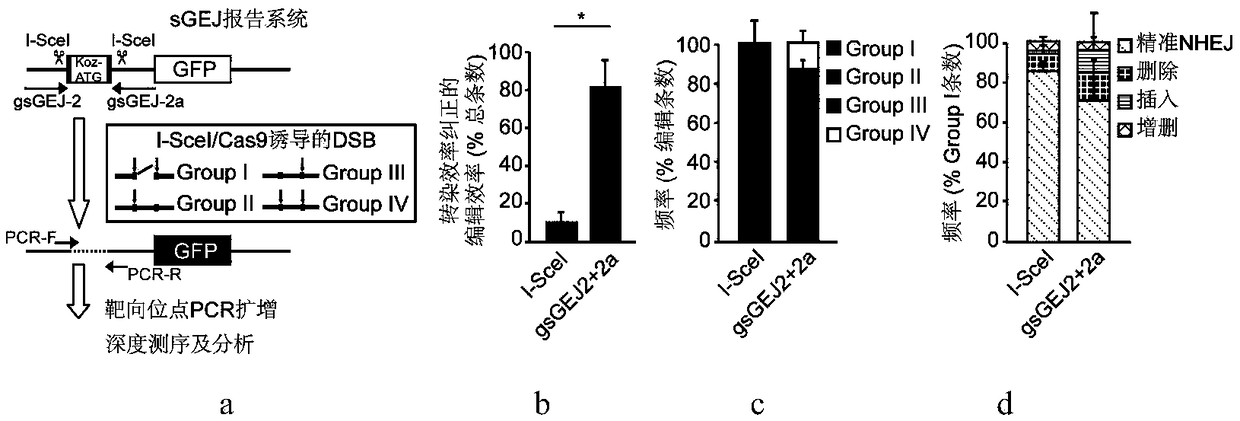
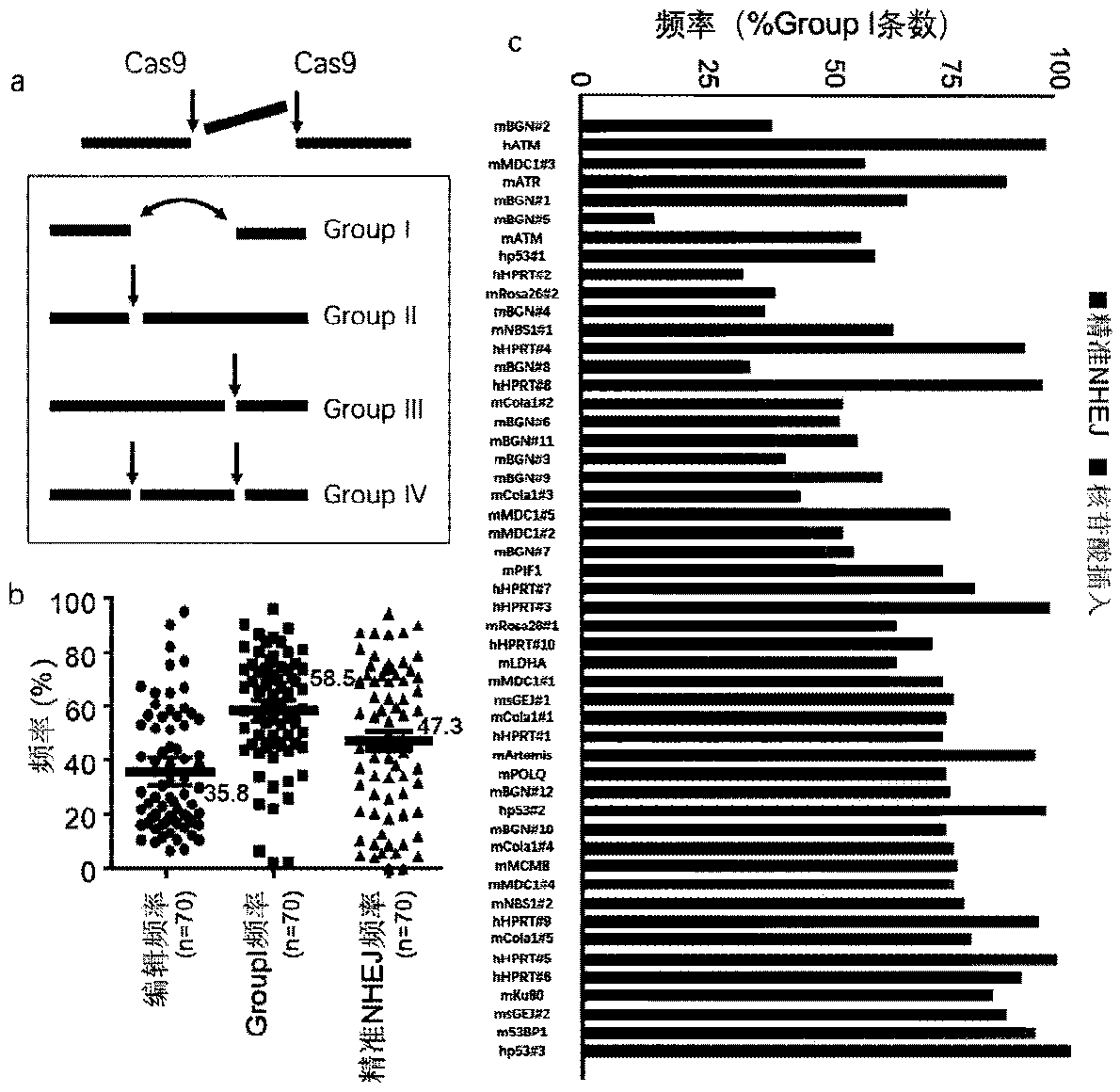
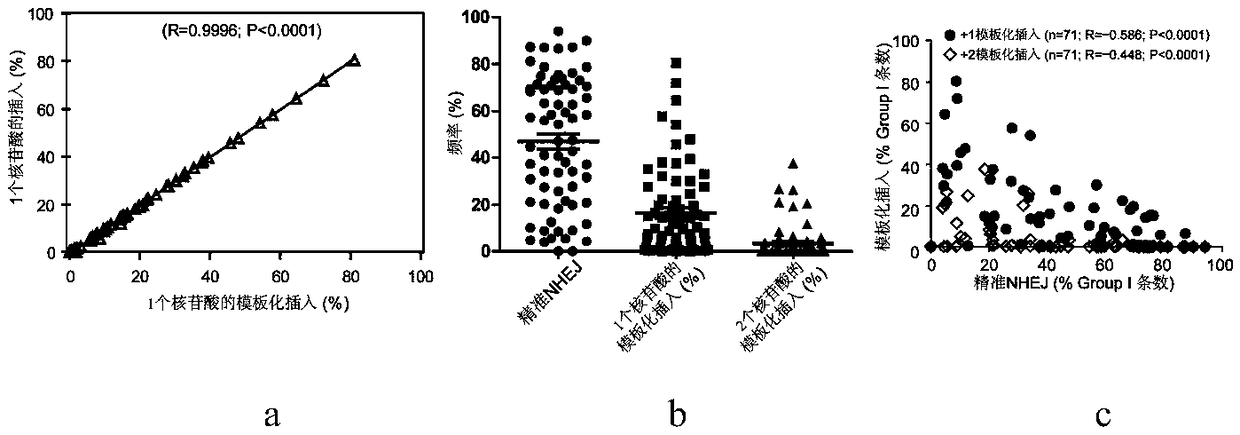
[0050] In recent years, gene editing technology using a single sgRNA to guide Cas9 has been widely used, especially gene knockout based on frameshift mutations. This technology uses non-precise NHEJ repair to generate additions and deletions, of which only some additions and deletions (theoretically two-thirds) will result in a frameshift mutation of a single allele and knock out one of the alleles, but the addition and deletion base sequences are mixed and long and short. Different, NHEJ products are highly heterogeneous, and the efficiency of simultaneous biallelic knockout is low. At the same time, this application also ignores an important problem, that is, because the ends of the DSB generated by Cas9 cleavage can be directly connected, NHEJ repair may be mainly accurate, and the efficienc...
Embodiment 2
[0226] Example 2: Knock out the MDC1 gene
[0227] In order to use paired Cas9-sgRNA technology to knock out mouse MDC1 (gene ID: 240087) gene, we designed three pairs of sgRNA (mMDC1-1: ACAGATCATGGAAAGCACCC; mMDC1-2: AGCATCCCAGTCAATCACCT; mMDC1) on exon 2 of MDC1 gene. -3: AACTATCCAGTGGCTCCTT), a single sgRNA and a paired sgRNA were respectively transfected into mouse embryonic stem cells. After 3 days, PCR amplification and DNA electrophoresis analysis found that the paired sgRNA2 / sgRNA3 (C / W, 52bp) had high editing efficiency ( Figure 15 In a), deep sequencing also shows that the efficiency of precise ligation is about 50%, and the frequency of templated insertion is about 5% ( Figure 15 In b). Therefore, we chose this pair of sgRNA to guide Cas9 to knock out the MDC1 gene, and select a monoclonal cell line from the cells edited by the paired Cas9-sgRNA. Specifically, the selected paired Cas9-sgRNA expression plasmid is transfected into mouse embryonic stem cells, the transf...
Embodiment 3
[0232] Example 3: Deleting gene fragments encoding 53BP1 domain with precise whole code
[0233] The paired sgRNA technology can control the length of excision fragments and can be used for precise whole-code excision, which can realize the functional study of specific gene fragments without generating frameshift mutations, especially for the study of some lethal gene domain fragments. In order to verify this application, we selected the 53BP1 gene (Gene ID: 27223) as the research object, targeting the key domains of 53BP1, Tudor and OD, which are necessary for the recruitment of 53BP1 protein to the DSB injury site ( Figure 16 In a). We designed paired sgRNAs for these two domains (Tudor domains: GGGAAGATCACCCGAGATGT and GGGTACGAATGTGACGTGCT; OD domains: GAGTGTACGGACTTCTCGAA and TTATGTGGATGGGACAGAAG). The length of the excised DNA fragments between these two paired sgRNAs is 3n bases. In this way, in gene editing, the two broken ends can be connected by precise NHEJ to achieve ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com