Single-cell methylation sequencing technology and application thereof
A single-cell, methylation technology, applied in the field of single-cell methylation sequencing technology, can solve the problems of low mapping rate of sequencing data, data waste, time-consuming and labor-intensive, etc., and reduce the probability of random primers and adapter sequences. , The effect of high Mapping rate and short manual operation time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
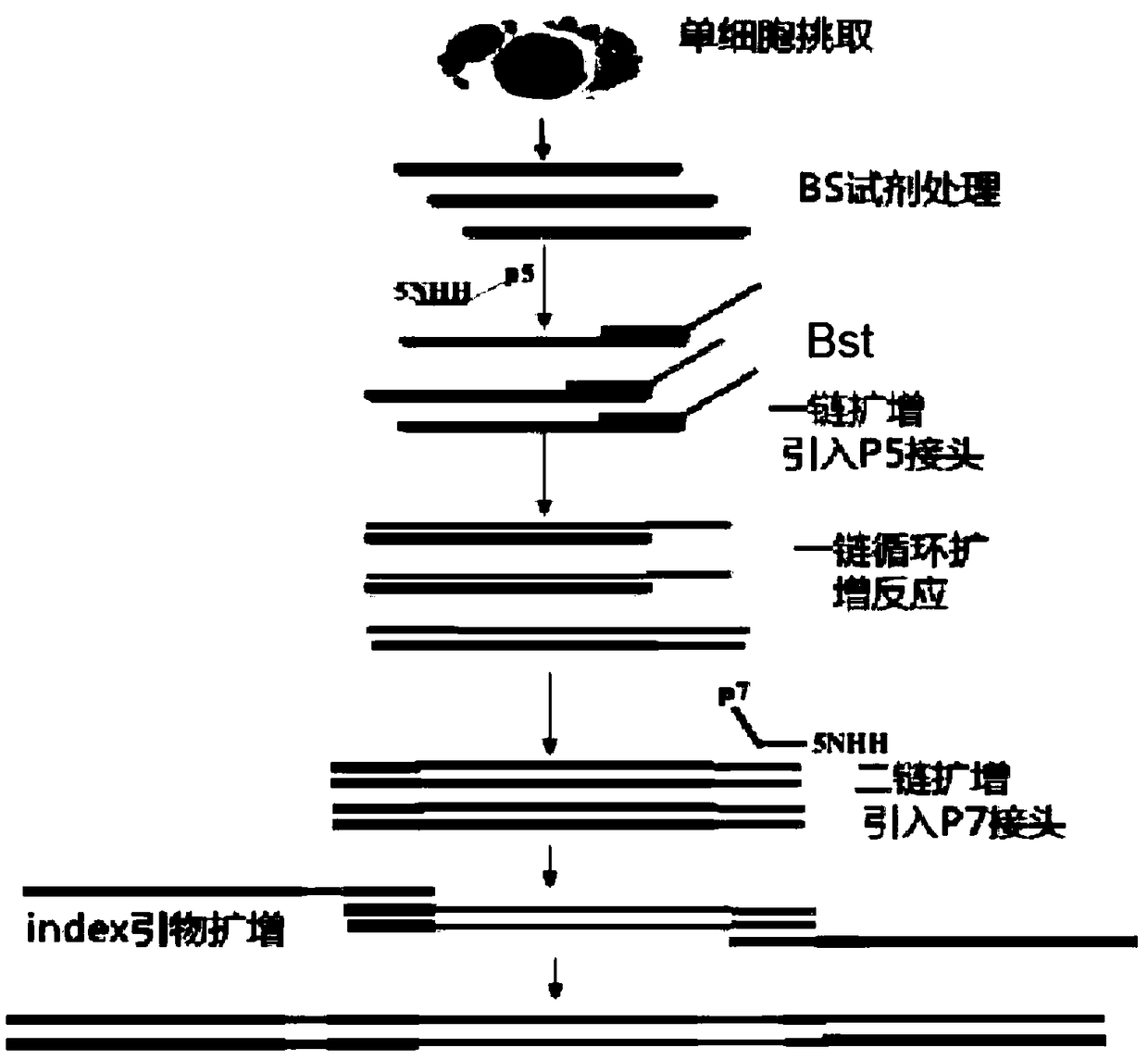
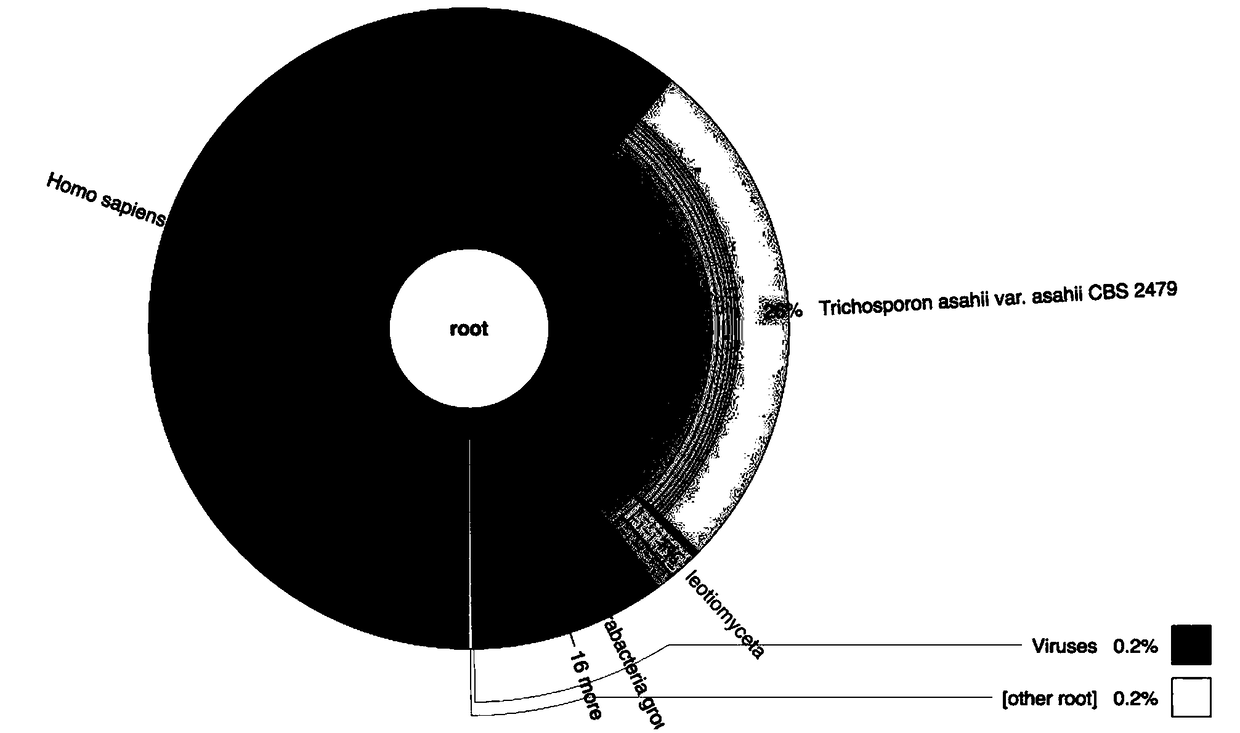
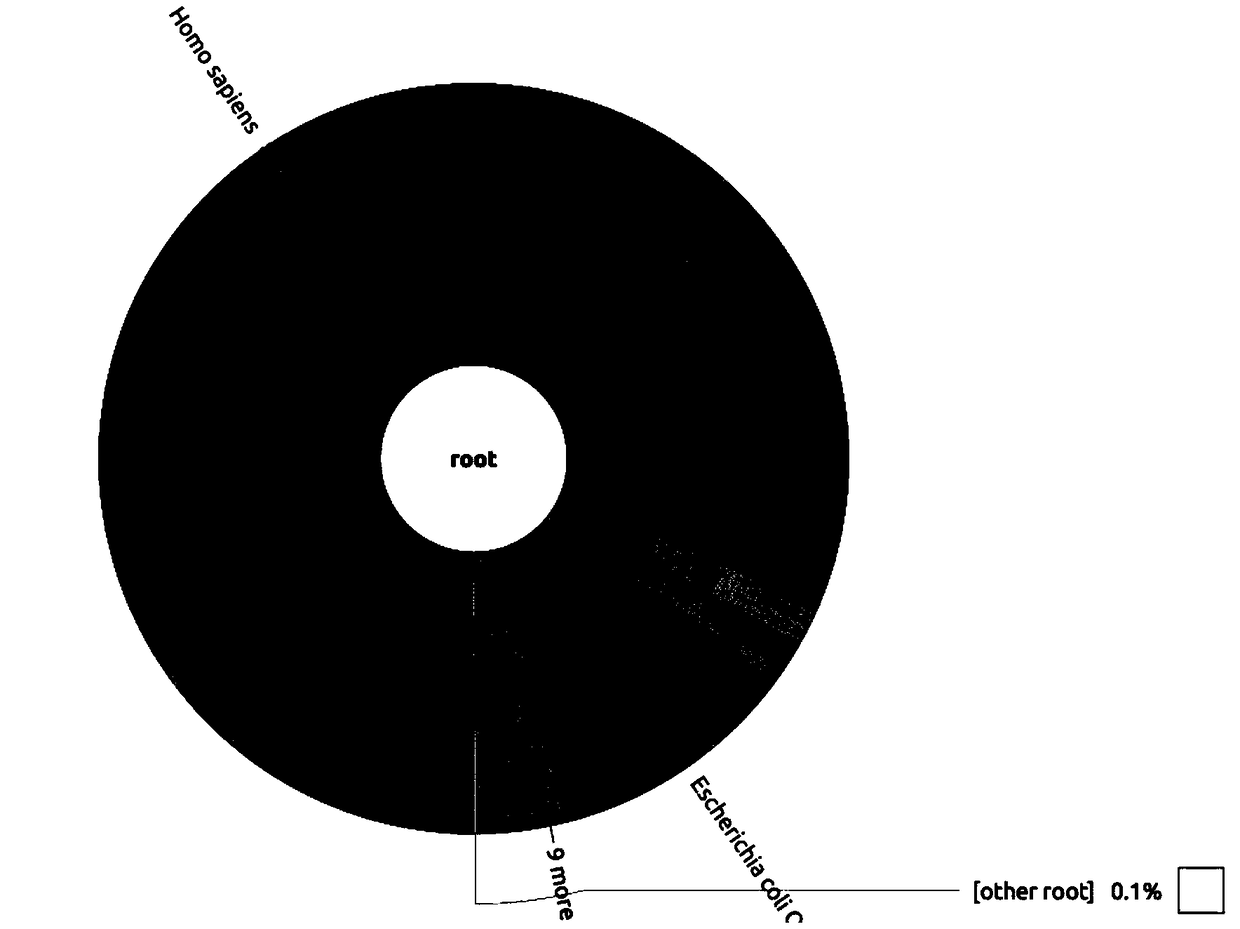
Image
Examples
Embodiment 1
[0040] Example 1. Establishment of single-cell methylation sequencing method
[0041] For the process of building a database, see figure 1 . In this embodiment, A549 cells are used as an example for operation.
[0042] 1. Single cell picking
[0043] 1. Take 2.5 μL single cell lysate buffer (BufferRLT Plus, QIAGEN, catalog number: 1053393), put it in a 0.2 mL centrifuge tube, centrifuge briefly (≤600 g, ~5 sec), and set aside.
[0044] 2. Pick a single A549 cell, place it in the lysate from step 1, and lyse it at room temperature for 10 minutes.
[0045] 2. Bisulfite conversion
[0046] Any bisulfite conversion kit can be used for bisulfite conversion.
[0047] In this example, the EZ DNAMethylation-Lightning kit (ZYMO research) was used for bisulfite conversion. The specific steps were as follows:
[0048] (1) Incorporate a certain amount of Lambda DNA into each cell, add H 2 O to a final volume of 10 μL.
[0049] (2) Add 600μL M-Binding Buffer to Zymo-Spin TM IC Co...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com