Multi-layer network model construction method and application with cancer-related SNP, gene, miRNA and protein interaction
A multi-layer network, construction method technology, applied in the fields of bioinformatics, biological systems, instruments, etc., can solve the problems of inability to obtain correlations and insufficient detection of genetic variants
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
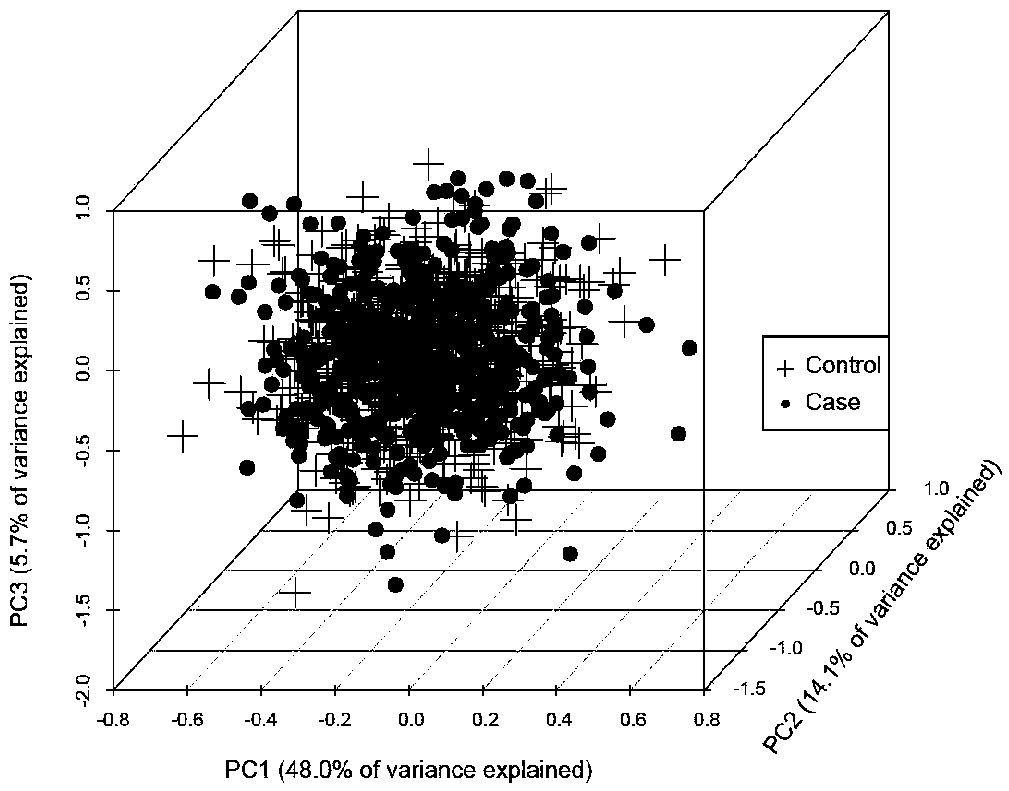
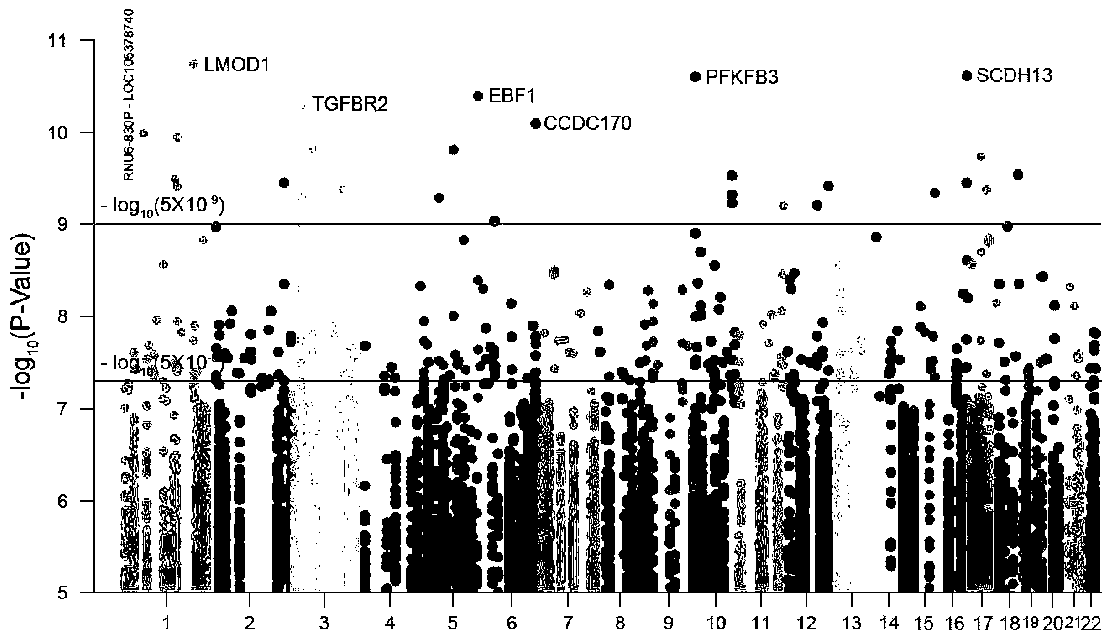
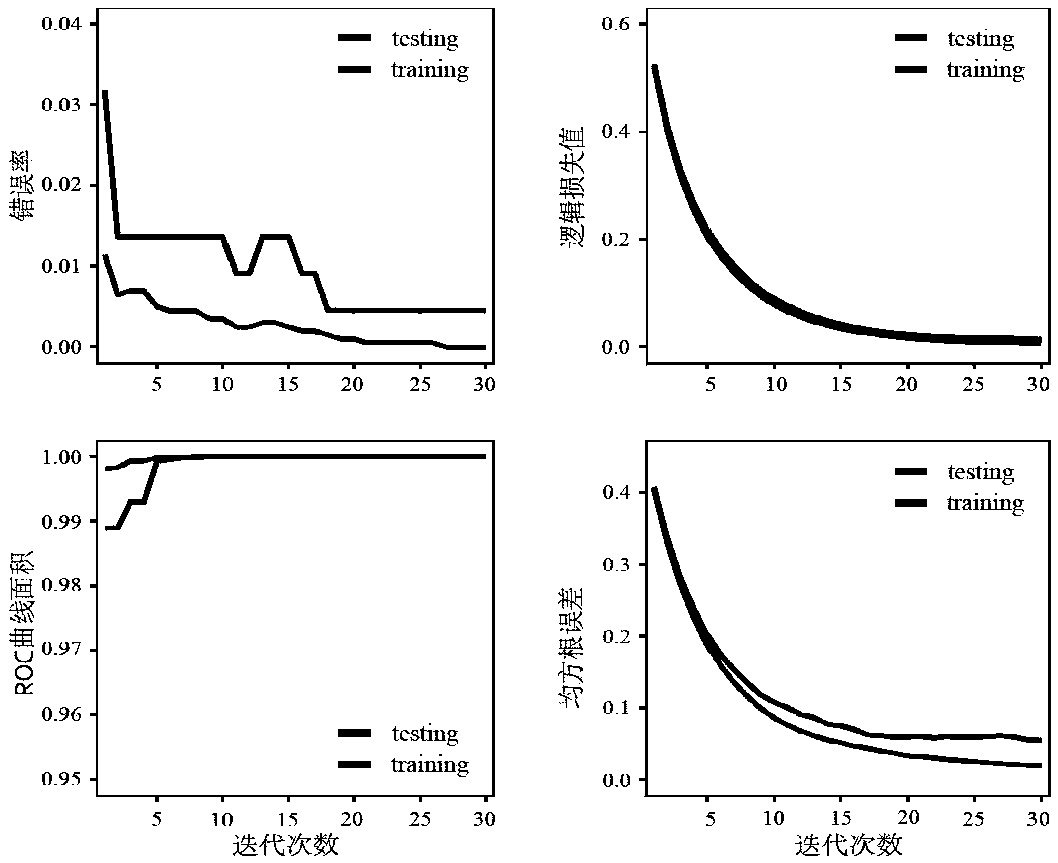
[0085] The genome-wide VCF data of breast cancer comes from the breast cancer laboratory database of the University of Cambridge. The downloaded data has a total of 1011 cases, including 640 breast cancer patients and 371 healthy controls. The breast cancer expression level data used to construct the network comes from the TCGA cancer database. Download RNA-Sequencing (hereinafter referred to as gene expression data), miRNASeq (hereinafter referred to as miRNA expression data) and protein expression data of breast cancer patients. For details, see Table 1.
[0086] Table 1 Tumor marker data description table
[0087]
[0088] The amount of gene expression data is 1214×24991 (the number of tumor tissue samples is 1101, the number of normal tissue samples is 113; 24991 is the gene type); the amount of miRNA expression data is 1200×1882 (the number of tumor tissue samples is 1096, and the number of normal tissue samples The number is 104; 1882 are miRNA types); the amount of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com