T-CELL RECEPTORS WHICH RECOGNISE FRAMESHIFT MUTANTS OF TGFbetaRII
A technology of cell receptors and immune effector cells, applied to genetically modified cells, animal cells, receptors/cell surface antigens/cell surface determinants, etc., can solve problems such as the impossibility of predicting the functional sequence of the TCR chain
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0241] Materials and methods:
[0242] Cell Lines, Media and Reagents
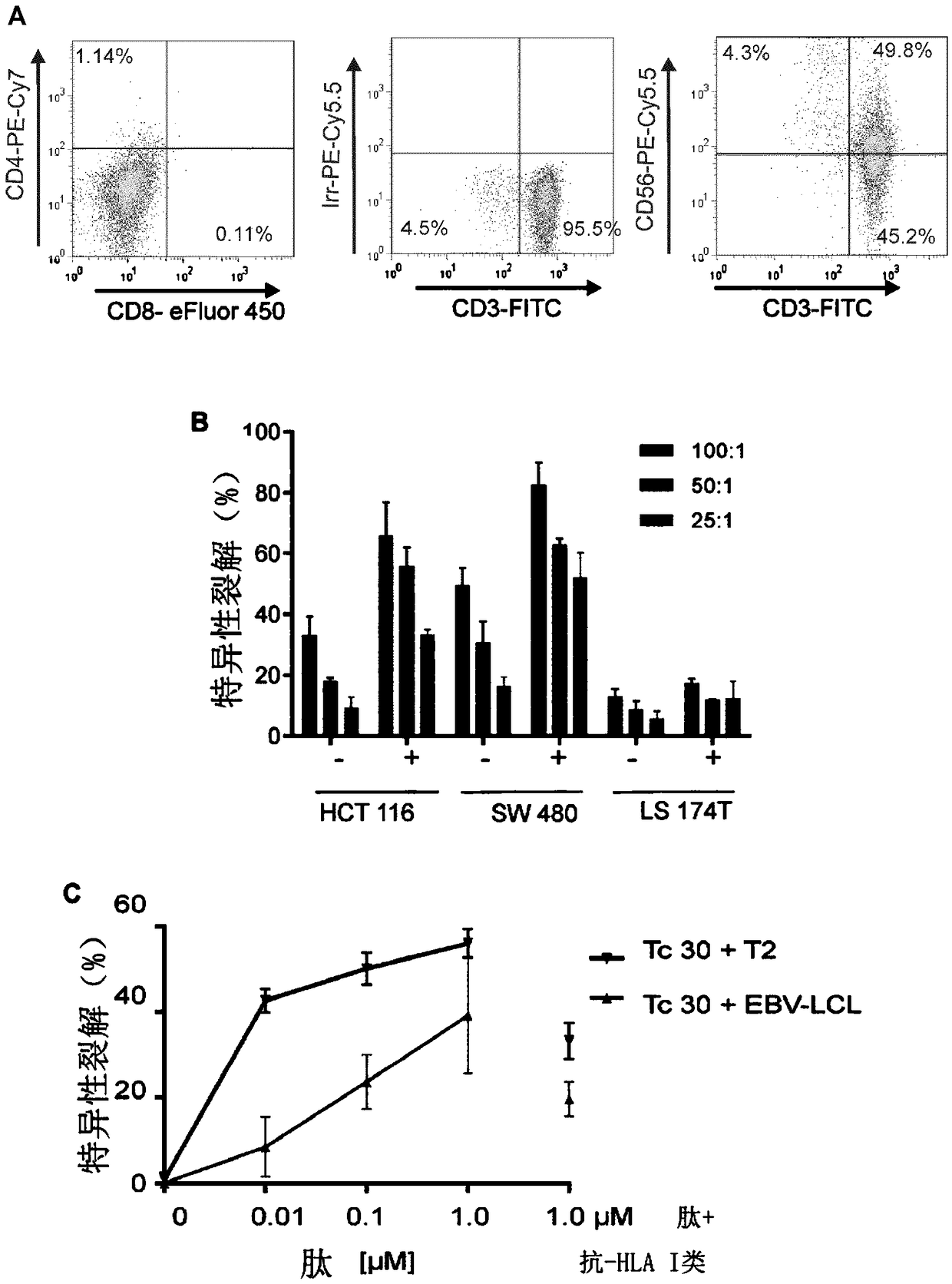
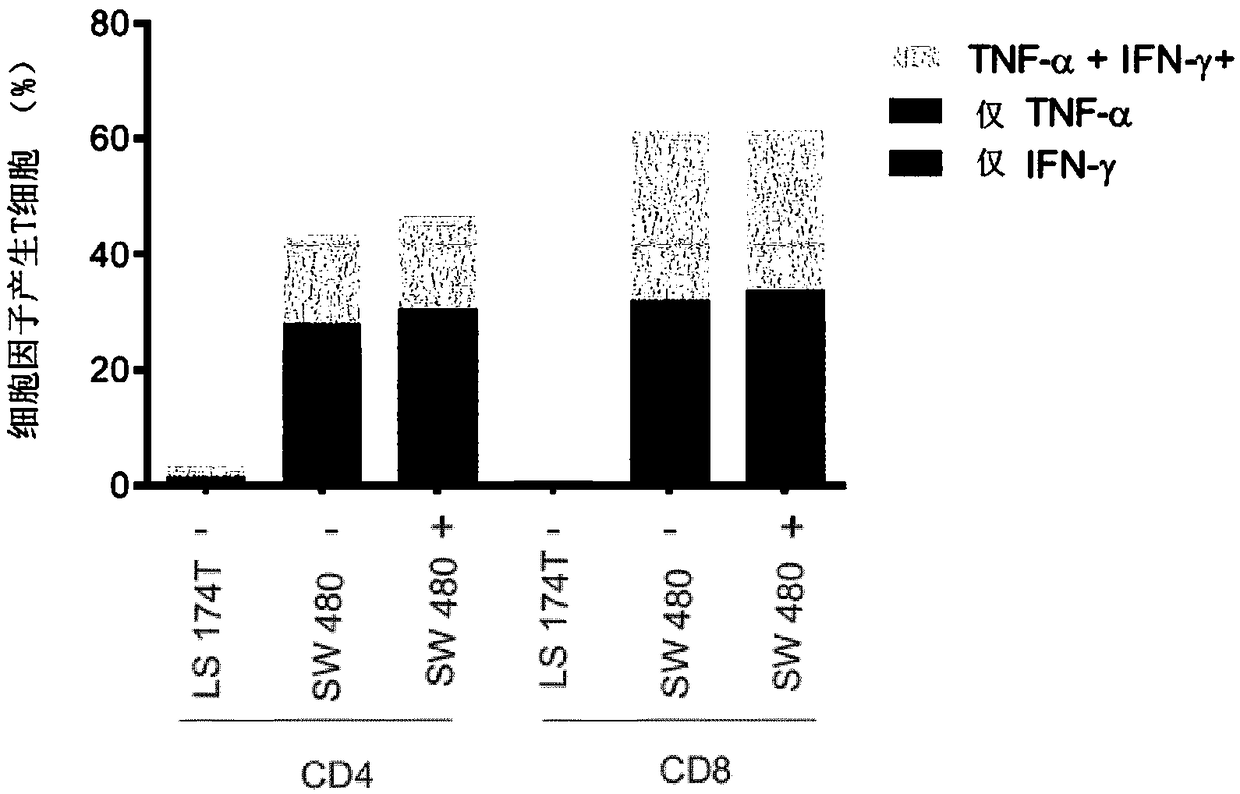
[0243] TGFβRII frameshift mutation-responsive, HLA-A2-restricted CTL (cytotoxic T lymphocyte) clones were isolated from the blood of MSI+ colon cancer patients and cloned by limiting dilution. The patient was vaccinated with the 23-mer TGFβRII(-1A) frameshift peptide (peptide having SEQ ID NO: 49). The clinical trial was approved by the Norwegian Medicines Agency, the Southern Medical Research Ethics Committee and the Hospital Review Board. Treatment was carried out in accordance with the Declaration of Helsinki of the World Medical Association. Informed consent was obtained from the patient. An autologous Epstein Barr virus transformed lymphoblastoid cell line (EBV-LCL) was generated by transformation of B cells from a donor. Antigen processing-deficient T2 cell lines are used as T cell targets in flow cytometry and cytotoxicity assays. Colon cancer cell lines HCT116, SW480 and LS174T and human emb...
Embodiment 2
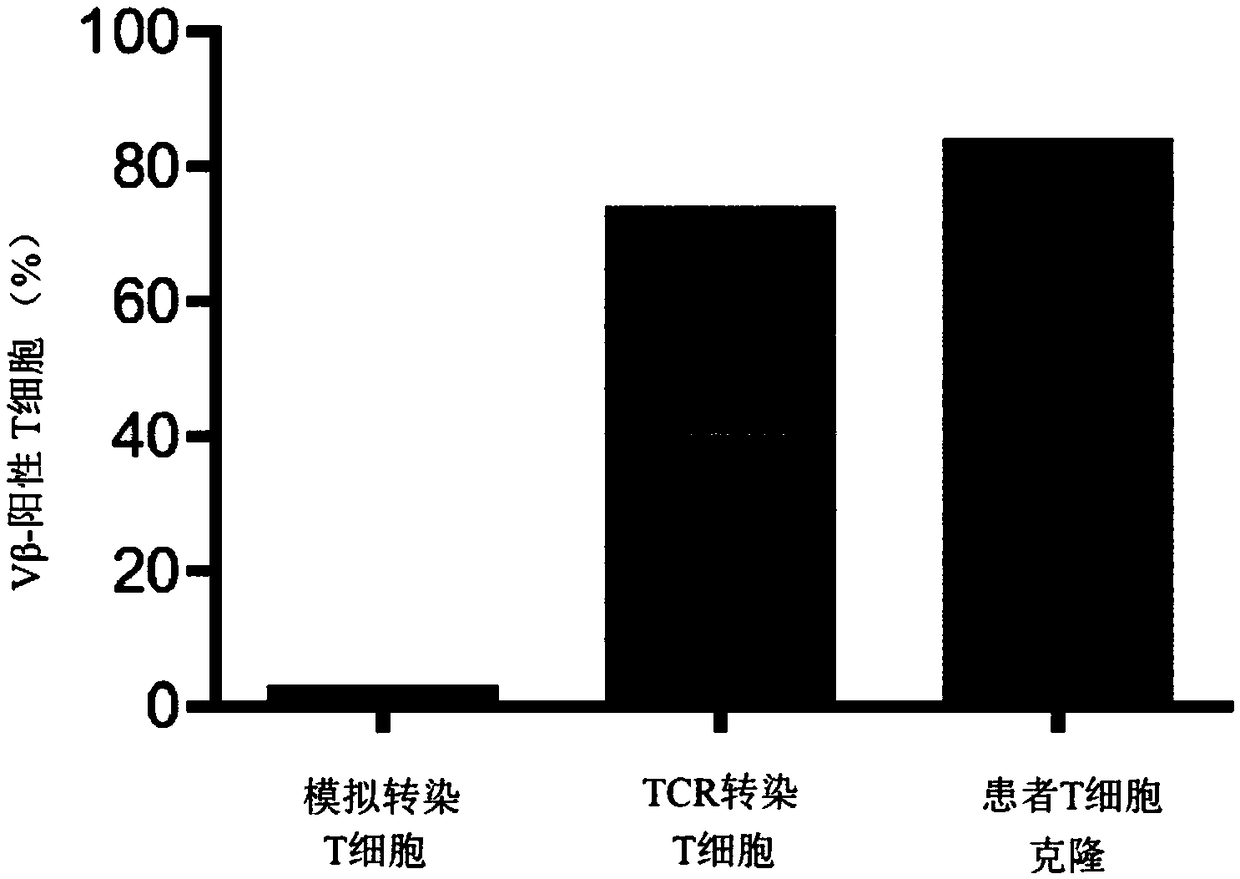
[0282] To study CD4 transduced with Radium-1 TCR + and CD8 + T cells kill target cells, which are stably transduced to express luciferase. Two sets of target cells were used: the HCT116 cell line and the Granta cell line. HCT116 cells are described above; the Granta cell line is a human B-cell lymphoma cell line. Changes in bioluminescence were used to measure changes in the number of target cells during culture of Radium-1-transduced T cells, representing killing of target cells by T cells.
[0283] Luciferase-transduced target cells were co-cultured with effector T cells on effectors to a target (E:T) ratio of 30:1 and bioluminescence was measured. Cells were co-cultured for 24 hours and bioluminescence was measured at 1, 2, 3, 4, 5, 8, 11, 20, 21, 22, 23 and 24 hours. Effector T cells were co-cultured with Granta cells with and without exogenous peptide p621 (SEQ ID NO:55) comprising the sequence of SEQ ID NO:1.
[0284] purified CD4 + T cells and purified CD8 transdu...
Embodiment 3
[0291] The soluble His-tagged Radium-1 TCR encoded by the scTCR of SEQ ID NO: 69 was expressed in HEK cells. Supernatants of cells expressing HEK were isolated. SupT1 cells (HLA-A2 negative cell line) were transduced to express HLA-A2 fused to a non-specific irrelevant peptide not recognized by Radium-1 or a TGFβRII frameshift peptide. Transduced cells were incubated with soluble Radium-1 TCR for 30 minutes at room temperature; non-transduced cells were also incubated with soluble TCR as an additional negative control. After incubation, cells were washed and then stained with allophycocyanin (APC) to identify soluble TCR binding. Staining was performed using a primary mouse anti-His antibody followed by a secondary APC-conjugated anti-mouse IgG antibody. The stained cells were then analyzed by flow cytometry ( Figure 12 ).
[0292] like Figure 12 As shown in A and 12B, essentially no staining of the negative control was observed, demonstrating that soluble Radium-1 TCR ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com