Recombined bursicon protein for facilitating increase of caridina antimicrobial peptide and application thereof
An antibacterial peptide, rice shrimp technology, applied in the biological field, can solve the problems of shrimp and crab anorexia, muscle white turbidity, body surface ulcer, etc., and achieve the effect of reducing mortality and improving antibacterial ability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
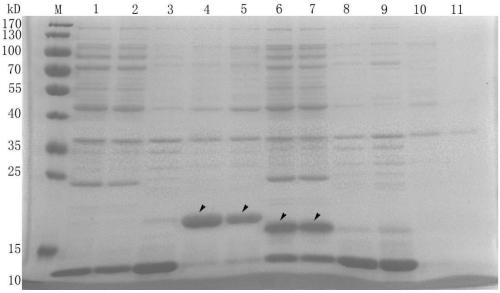
Image
Examples
Embodiment Construction
[0032] Zhejiang Rice Shrimp is purchased at the Zhejiang Aquatic Products Market. Its body length is about 1.5-2.5cm, and its weight is about 50-100mg each. It is healthy and energetic. The rice shrimps are raised in the laboratory after they are purchased. The water is aerated and the temperature is about 26-28°C. Shrimp food was fed twice a day, and fresh aeration water was replaced every two days.
[0033] Short distance: the time is 3s, 1000rpm.
[0034] bursicon-α has the DNA sequence described in SEQUENCE LISTING NO.1.
[0035] Bursa protein has the amino acid sequence described in SEQUENCE LISTING NO.2.
[0036] bursicon-β has the DNA sequence described in SEQUENCE LISTING NO.3.
[0037] Bursβ protein has the amino acid sequence described in SEQUENCE LISTING NO.4.
[0038] The technical solutions of the present invention will be further described below in conjunction with specific embodiments.
[0039] Step 1, Trizol method to extract total RNA
[0040] Take a 1.5m...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Relative molecular weight | aaaaa | aaaaa |
| Relative molecular weight | aaaaa | aaaaa |
| Relative molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com