Recombinant nucleotide fragments Rec78801 and Rec78802, detection primer and application thereof
A nucleotide and fragment technology, which can be used in recombinant DNA technology, DNA/RNA fragments, and microorganism determination/inspection, etc., and can solve the problems of low efficiency and long breeding cycle.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0098] Breeding of Individual Plants Introduced with Bph14 and Bph15 Fragments of Resistance to Brown Planthopper
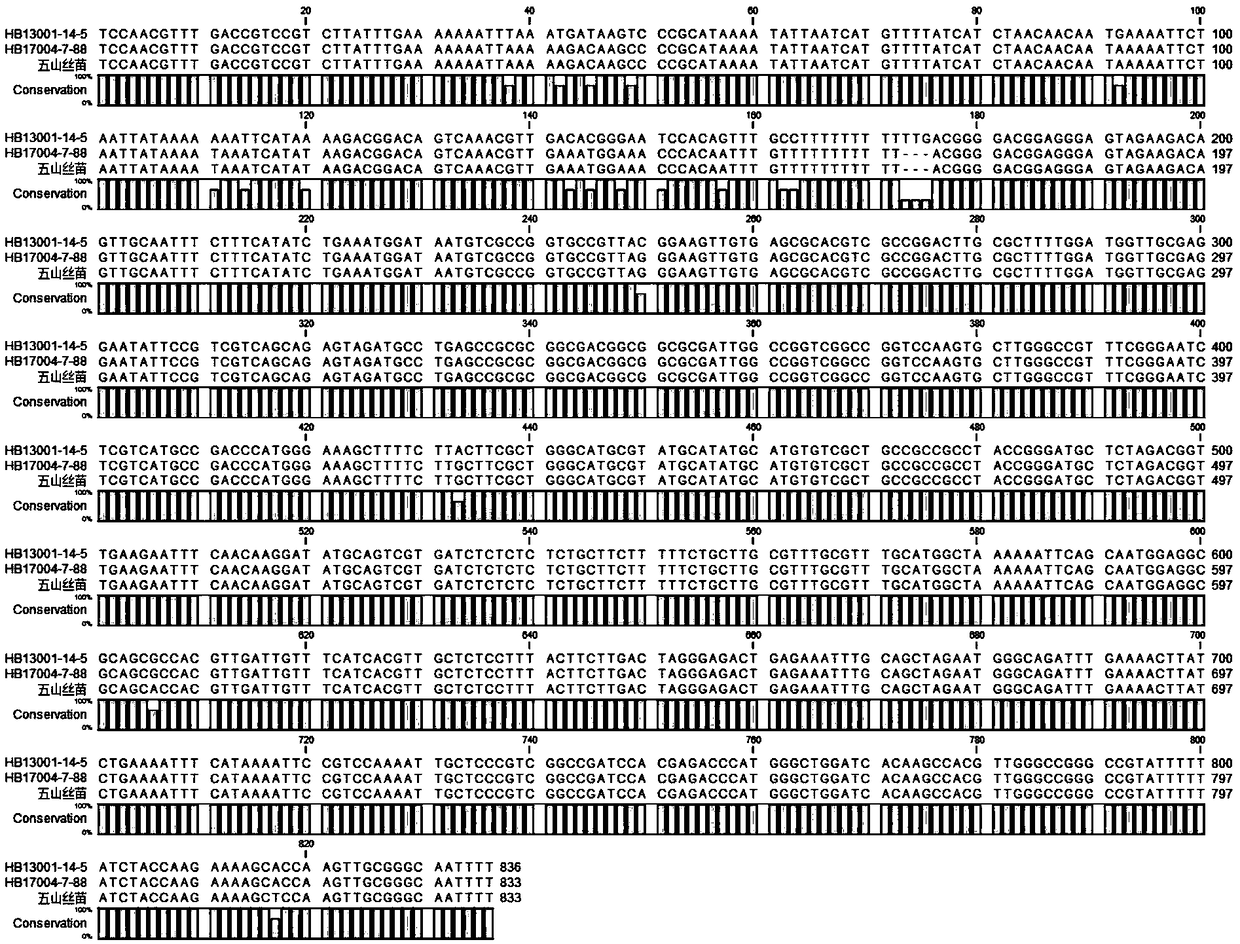
[0099] The rice materials used in this example are "Wushan Simiao" and "HB13001-14-5".
[0100] Among them, "Wushan silk seedlings" were provided by Hefei Quanyin Hi-Tech Seed Industry Co., Ltd.;
[0101] "HB13001-14-5" is a rice material carrying brown planthopper resistance genes Bph14 and Bph15 independently selected and bred by Huazhong Agricultural University.
[0102] Import the genomic DNA fragments where the Bph14 and Bph15 genes in "HB13001-14-5" are located into "Wushan Simiao", the specific process is as follows:
[0103] (1) Screening of foreground selection markers: by searching the SCI documents related to the two brown planthopper resistance genes involved in the present invention, molecular markers closely linked to the target gene were obtained, and good polymorphism between the two parents was obtained by screening Molecular markers of 76-2 an...
Embodiment 2
[0114] Determination of the Homologous Recombination Fragment after Introducing the Resistance Gene Fragment of Brown Planthopper
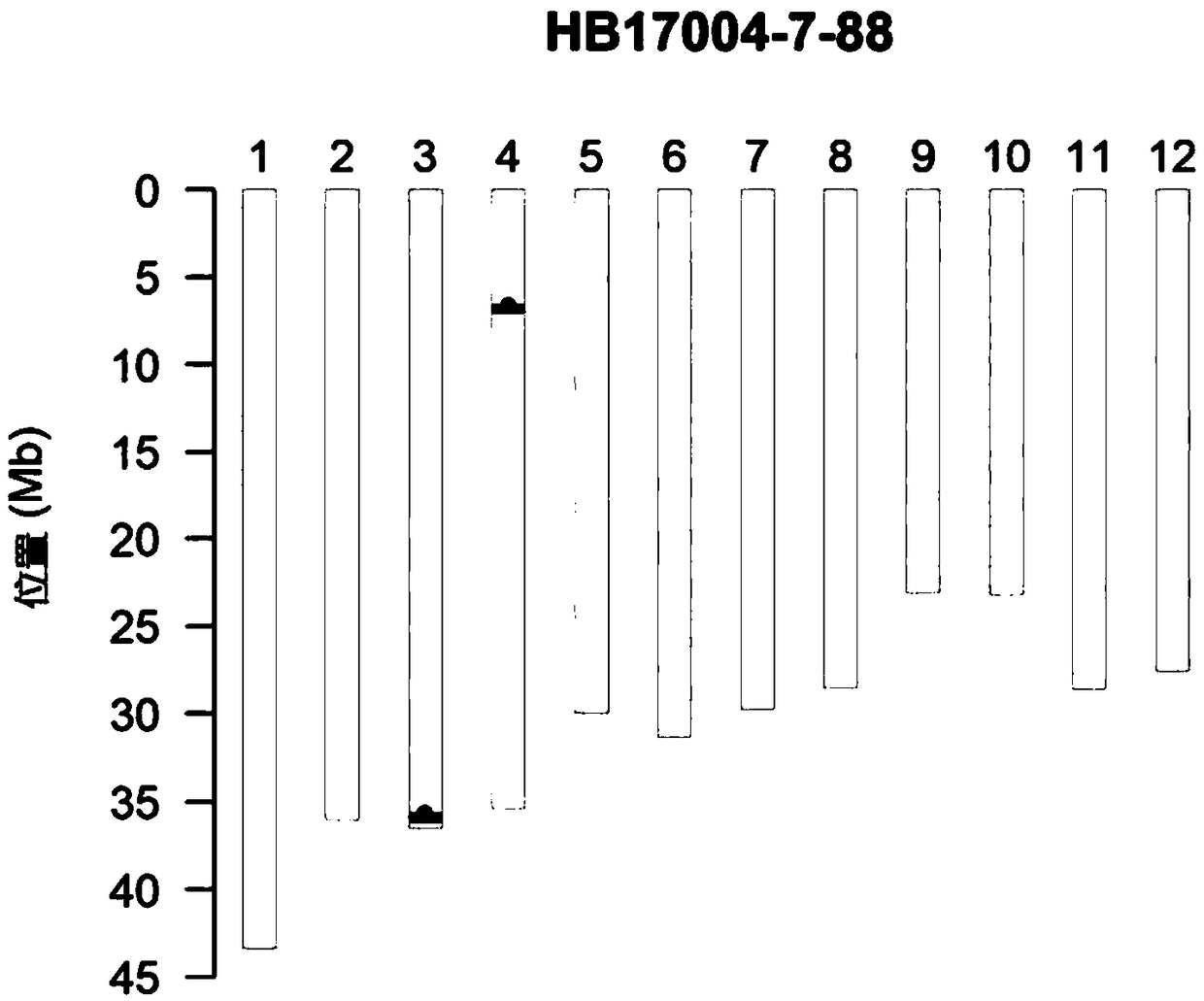
[0115]In order to determine the size of the imported N. lugens resistance gene fragment, the homologous recombination regions on both sides of the two N. lugens resistance gene fragments introduced into the 'Wushan Simiao' background were mapped and sequenced. First, according to the GSR40K chip detection results, the upstream homologous recombination region of Bph14 was located in the approximately 9.1kb region between SNP markers F0335655549TG and F0335664621CT (chromosome 3 35655549bp-35664621bp); the downstream homologous recombination region of Bph15 was located in the SNP marker F0407000448AG About 870bp region (7000448bp-7001318bp) between F0407001318GC and F0407001318GC.
[0116] In order to further narrow the upstream homologous recombination interval of Bph14, on the basis of the chip results, the DNA sequence of the corresponding segmen...
Embodiment 3
[0123] Resistance Identification
[0124] In order to identify the resistance effect of 'Wushan Simiao' after the introduction of N. lugens resistance genome fragments (target individual plants), "HB17004-7-88", "Wushan Simiao", "HB13001-14-5", and "TN1" and "B5" for indoor planting and artificial inoculation of brown planthopper resistance identification, the specific methods are as follows:
[0125] Before the identification of artificial inoculation, the artificially captured N. lugens planthoppers were reared with TN1 (Tai Chang Native1) materials that did not carry any resistance gene. The specific method is: capture a certain number of wild brown planthoppers from the field and raise them on the TN1 seedlings planted in advance, and artificially raise them for multiple generations to increase the number of brown planthopper nymphs to meet the identification requirements.
[0126] Prepare 100 seeds each of 'HB17004-7-88', 'Wushan Simiao', 'HB13001-14-5', 'TN1' and 'B5', ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com