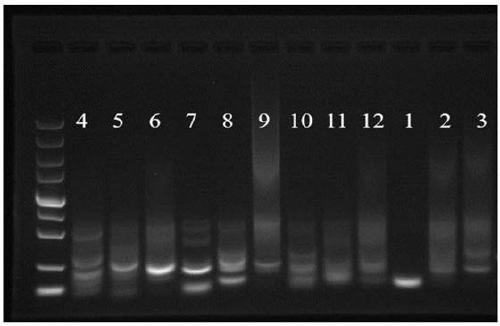
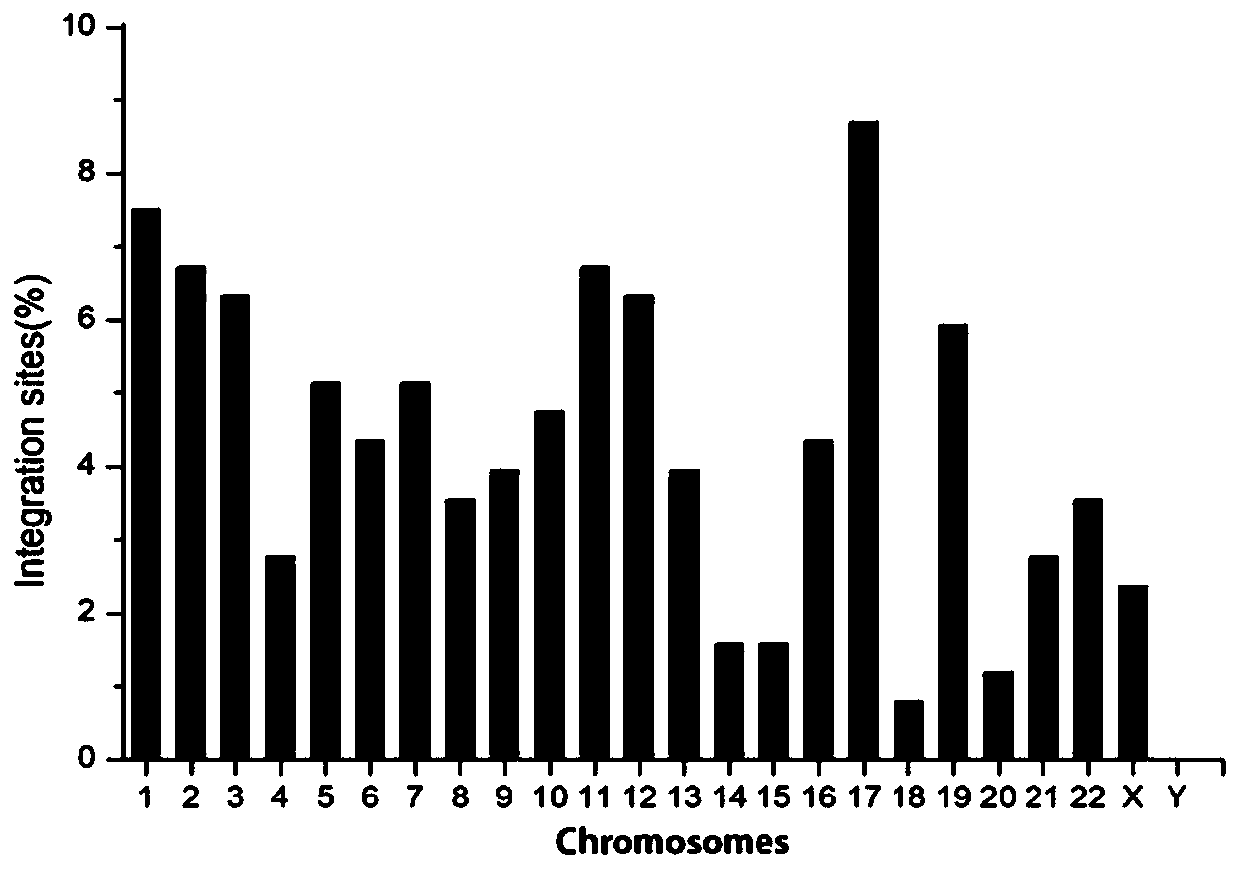
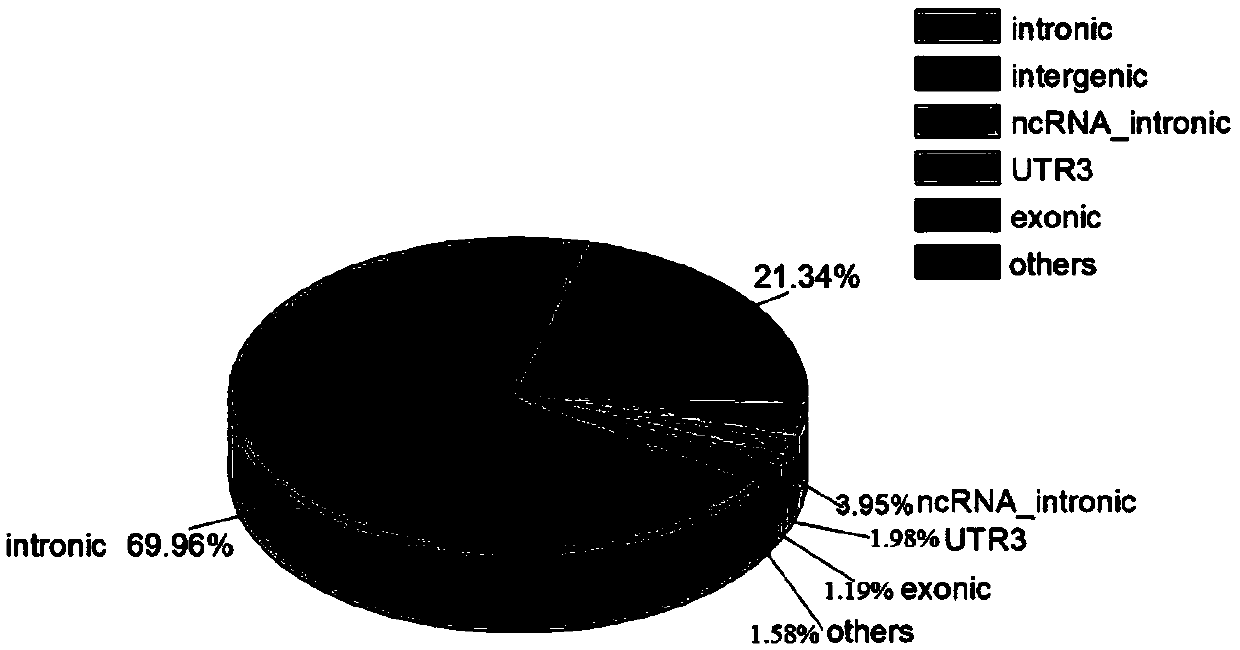
Method for analyzing an integration site of lentiviral vector in CAR-T cells, and primers
A technology of lentiviral vector and analysis method, which is applied to the analysis of the integration site of lentiviral vector in CAR-T cells. The primer sequence field of this method achieves the effect of good specificity, stability and high sensitivity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 Analysis of integration sites of lentiviral vectors in CAR-T cells
[0034] The primer sequences are shown in the table below:
[0035]
[0036] 1 Experimental method
[0037] 1.1 Extraction of total genome DNA of anti-BCMACAR-T cells
[0038] (1) Sample homogenate
[0039] Add 1ml DNAZOL Reagent to the CAR-T cell sample, and pipette 50 times, taking care to avoid air bubbles.
[0040] (2) DNA precipitation
[0041] Add 0.5ml of 100% ethanol to the above cell homogenate, turn it upside down 10 times, and let it stand at room temperature for 1-3 minutes, flocculents can be seen. Centrifuge at 15000g for 5min. Remove the supernatant with a pipette.
[0042] (3) DNA washing
[0043] Add 1ml of 75% ethanol, invert up and down 10 times, and centrifuge at 12000g for 1min. Carefully remove the supernatant with a pipette. repeat. Open the cap of the EP tube for 3-5 minutes, and let it dry in the air.
[0044] (4) DNA dissolution
[0045] According to th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com