FnCpf1 mediated in-vitro DNA editing kit
A kit and cpf1 technology, applied in the field of molecular biology, can solve the problems of complicated and time-consuming operation, limited flexible application, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0081] Example 1 DNA connection method based on CRISPR / Cpf1 system
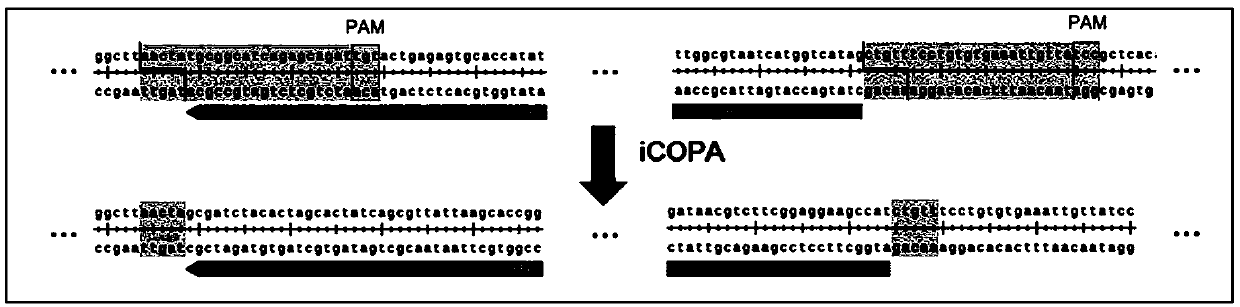
[0082] One, the test kit and method of iCOPA of the present invention
[0083] 1. Add sample
[0084]
[0085] Among them, the carrier and the linearized insert have a CrRNA recognition cutting region that guides the Cpf1 enzyme, and the CrRNA recognition cutting region is formed by connecting a PAM site and a spacer with a length of 23 bp;
[0086] CrRNA template DNA: composed of T7 promoter, DR, spacer sequence;
[0087] DR is 19 or 36nt, and the spacer sequence length is 23nt;
[0088] T7 promoter sequence: GAAATTAATACGACTCACTATAGG;
[0089] 19nt DR sequence: AATTTCTACTGTTGTAGAT;
[0090] 36nt DR sequence: GTCTAAGAACTTTAAATAATTTCTACTGTTGTAGAT;
[0091] The number of CrRNA template DNA is more than 1, and the spacer sequence is consistent with the 23nt length recognition region on the vector and the insert fragment respectively.
[0092] The transcription mixture (available from Tiangen Company) co...
Embodiment 2
[0133] Example 2 Screening experiment of the DNA connection method based on the CRISPR / Cpf1 system of the present invention
[0134] 1. Optimizing the conditions for in vitro shearing of Cpf1
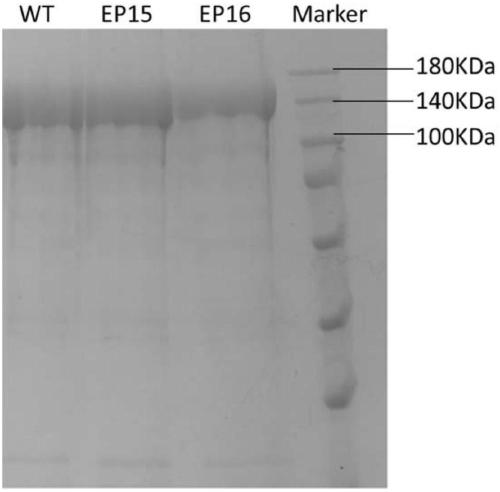
[0135] Wild-type FnCpf1 tends to cleave TTV PAM more than TTT). In order to obtain a suitable balance of reaction conditions corresponding to PAMTTN, to facilitate the universality and efficiency of subsequent toolbox experiment design. We first selected two groups of spacers (E-SP3: TTT PAM; E-SP15: TTA PAM) for in vitro enzyme activity testing. Linearized DNA T PAM 1 and T Red 1 were used as digestion substrates corresponding to spacer E-SP3 and spacer E-SP15, respectively. The sheared product fragments are 527bp and 707bp or 802bp and 2233bp, respectively.
[0136] According to the aforementioned system, perform the following screening to determine the parameters of the CRISPR / Cpf1 system:
[0137] 1. Test the effect of crRNA concentration on shearing efficiency, including 10nM...
experiment example 1
[0141] Experimental Example 1 Editing using the kit of the present invention: inserting fragments based on FnCpf1
[0142] The purpose of the experiment: add the promoter plac before the fluorescent protein expression gene mRFP, thereby activating the expression of the fluorescent protein mRFP;
[0143] 1. Select the spacer on the original vector, and design the corresponding spacer that can be applied to the promoter
[0144] (Table 1-1)
[0145] Table 1-1 The spacer applied to mRFP fluorescent protein activation
[0146]
[0147] 2. Obtain crRNA by PCR to prepare substrate template DNA
[0148] According to the requirements of the T7 transcription kit, design the corresponding primers (Table 1-2)
[0149] Table 1-2 is used to prepare substrate template DNA for crRNA activated by mRFP
[0150]
[0151] The obtained primers were used for PCR enrichment, and the enzyme used was Q5 high-fidelity enzyme (NEB Company, product number M0491). The specific configuration sy...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com