Genotype correction device and method
A genotype and Bayesian model technology, applied in genomics, instruments, biological systems, etc., can solve problems such as sequencing bias and mutation frequency deviation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
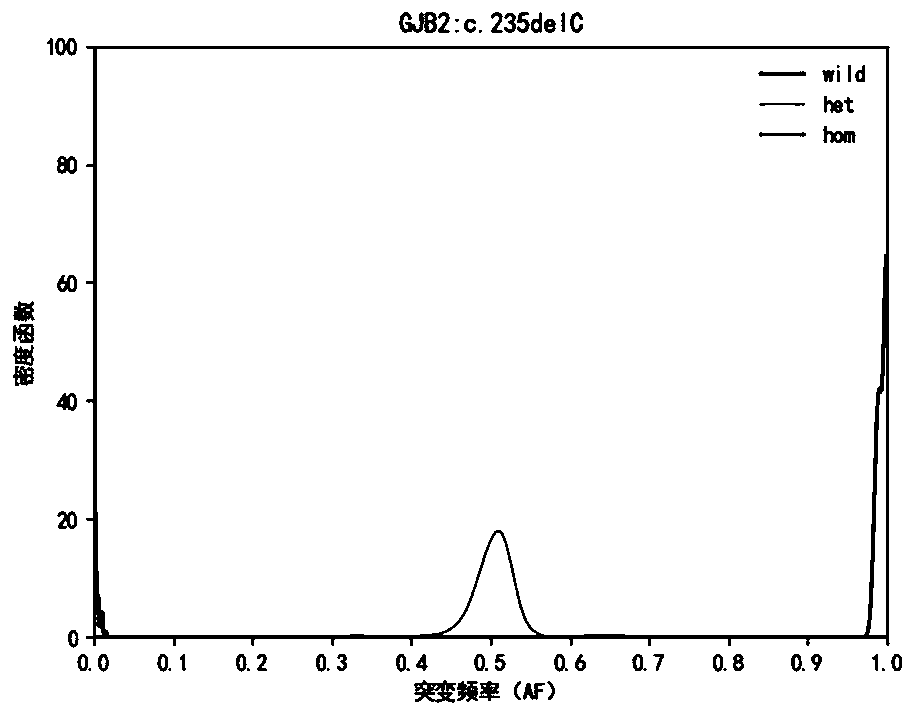
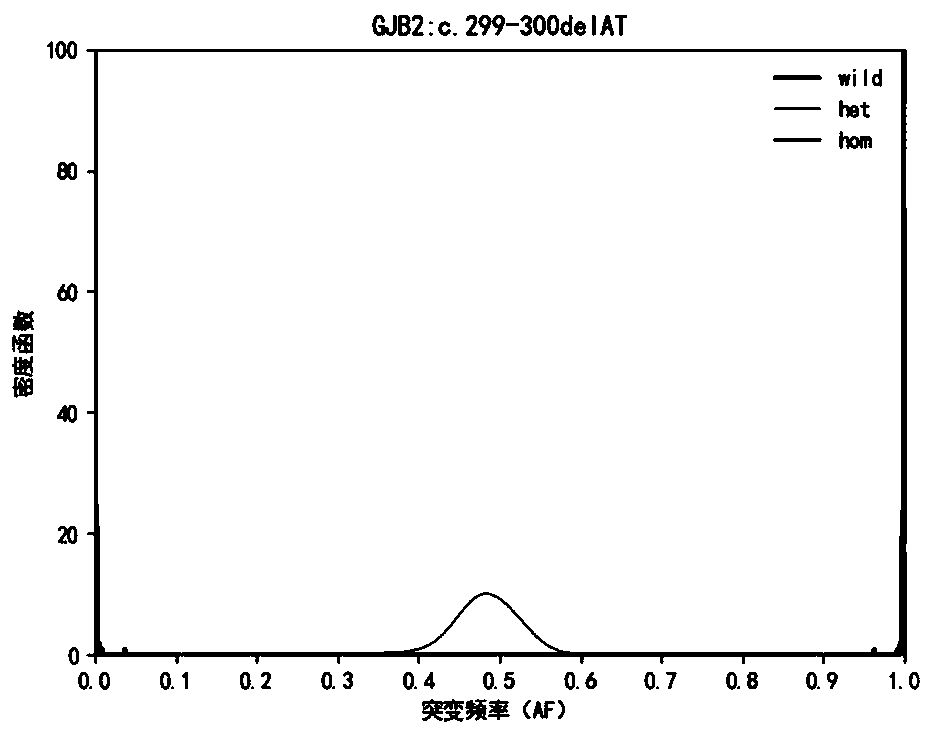
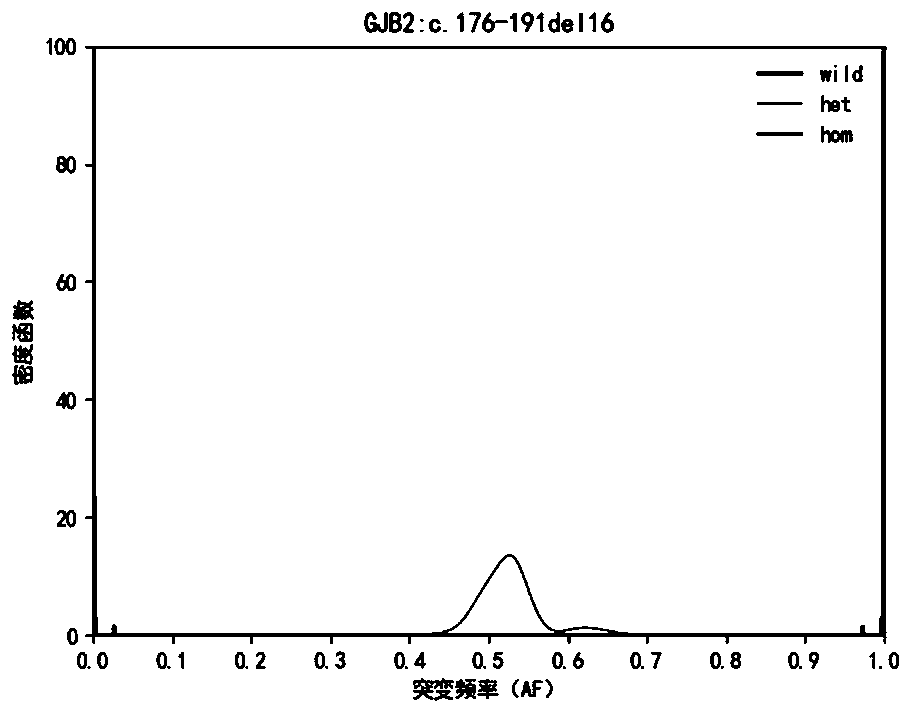
[0124] Hereditary deafness-related genes Detect 33 mutation sites of 5 common genes of hereditary deafness, design PCR primers for each site, amplify target region fragments by multiplex PCR technology, determine the genotype of each site It is determined based on the ratio of the number of mutant bases in the sequencing data of the site to be detected to the sequencing depth, that is, the mutation frequency (Allele Frequency, abbreviated as AF). For the detection results of high-throughput sequencing, due to fragment amplification, sequence Due to the sequencing bias caused by comparison, sequencing errors and other reasons, the measured value of the mutation frequency of the detection result will have a certain deviation from the true value.
[0125] This application builds a genotype frequency distribution model for each site based on Bayesian Models, and calculates the conditional probability value P(AF|GT) and the posterior probability value P( GT|AF), the genotype of the...
Embodiment 2
[0162] Example 2 result verification
[0163] The present invention carries out the establishment of threshold value by 730 samples, and the sensitivity and specificity of each point of each sample are all 1; Adopt 645 samples to verify the model threshold value formulated simultaneously, verification result is shown in Table 4:
[0164] Table 4 Validation results of the model threshold
[0165]
[0166]
[0167] It can be seen from Table 4 that the site sensitivity and specificity of all negative data are 1, which shows that the calculated sensitivity and specificity of each detection site are very good, which can meet the detection requirements of clinical samples, and the detection results are reliable.
Embodiment 3
[0169] Compared with the variation detection results without Bayesian combined kernel density estimation correction, the results are shown in Table 5
[0170] table 5
[0171]
[0172]
[0173]
[0174] It can be seen from Table 5 that there are occasional abnormalities in the detected variant genotypes of the variant detection results without Bayesian combined kernel density estimation correction, that is, the wild type is detected as a mutant type, and the heterozygous mutation is detected The sensitivity and specificity of genotype analysis by high-throughput sequencing can be improved after correction.
[0175] In summary, the present invention provides a device and method for genotype correction. The device establishes a statistical model for the detected data through a Bayesian model, constructs a priori distribution, determines the overall distribution information, and finally calculates the post- Posterior probability value P(GT|AF), through judging the relat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com