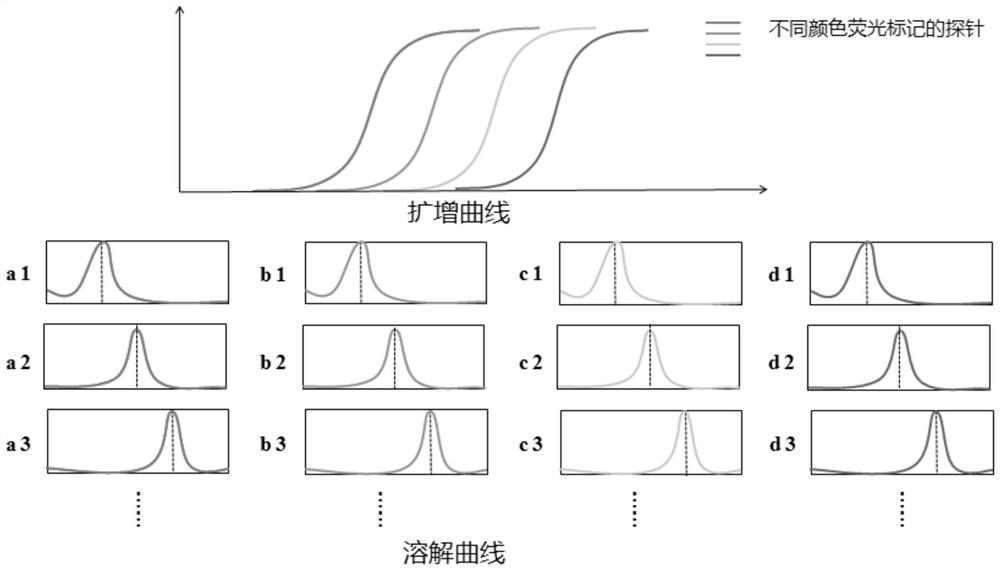
Fluorescent real-time detection reagent and method for simultaneously detecting multiple target genes
A target gene and target detection technology, applied in the field of fluorescent real-time detection reagents, can solve the problems of uncapping pollution, large product fragments, and reduced PCR efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0105] Example 1. Templates for amplifying 9 genes by single-tube multiplex qPCR
[0106] In this example, bacterial KPC (SEQ ID NO: 1), NDM (SEQ ID NO: 2), ACC (SEQ ID NO: 3), VIM (SEQ ID NO: 4), EBC (SEQ ID NO: 4) were obtained by gene synthesis NO: 5), CIT (SEQ ID NO: 6), OXA48 (SEQ ID NO: 7), GES (SEQ ID NO: 8) and DHA (SEQ ID NO: 9), a total of 9 drug resistance gene fragments were used as templates. The primer and probe sequences of each gene are shown below, wherein the probes of KPC, NDM and ACC are labeled with a Texas red fluorescent group at the 5' end and a BHQ2 quenching group at the 3' end; probe 5 of VIM, EBC and CIT The CY5 fluorophore was labeled at the ' end and the BHQ3 quenching group was labeled at the 3' end; the probes for GES, OXA48 and DHA were labeled with the HEX fluorophore at the 5' end and the BHQ2 quenching group at the 3' end.
[0107] KPC-F: CGGCAGCAGTTTGTTGATTG (SEQ ID NO: 10)
[0108] KPC-R: CAGACGACGGCATAGTCATTT (SEQ ID NO: 11)
[0109] K...
Embodiment 2
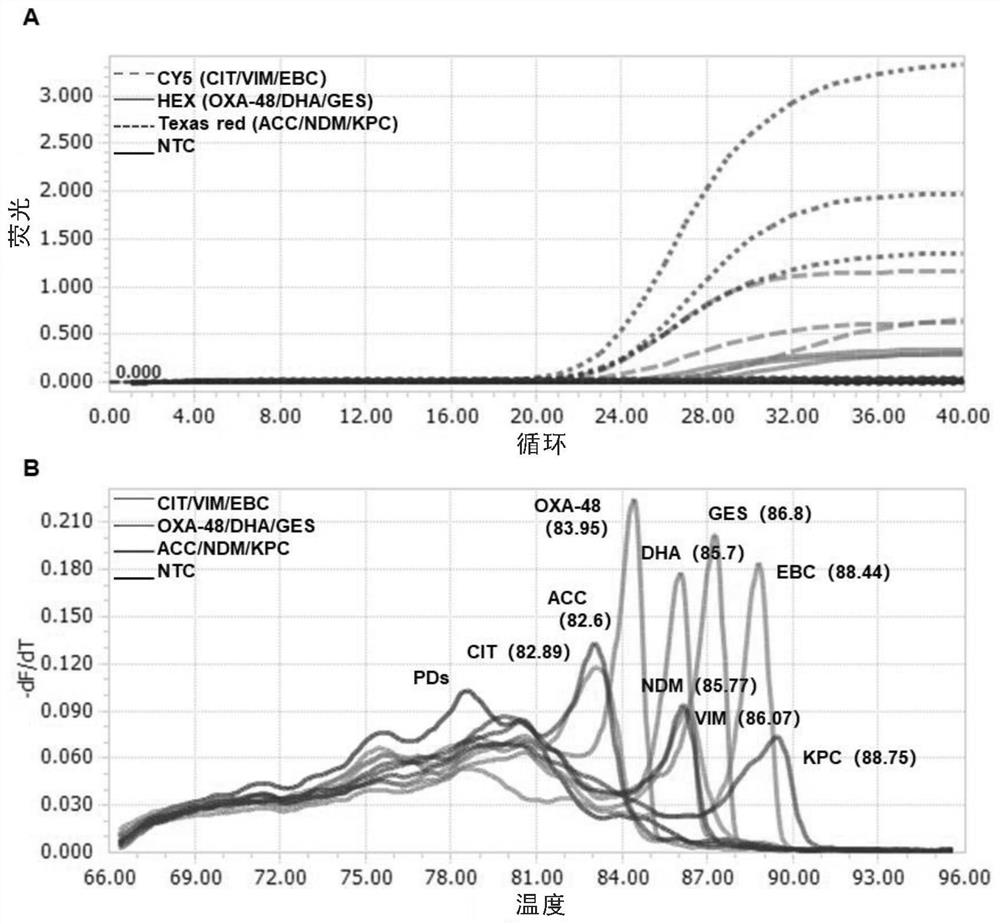
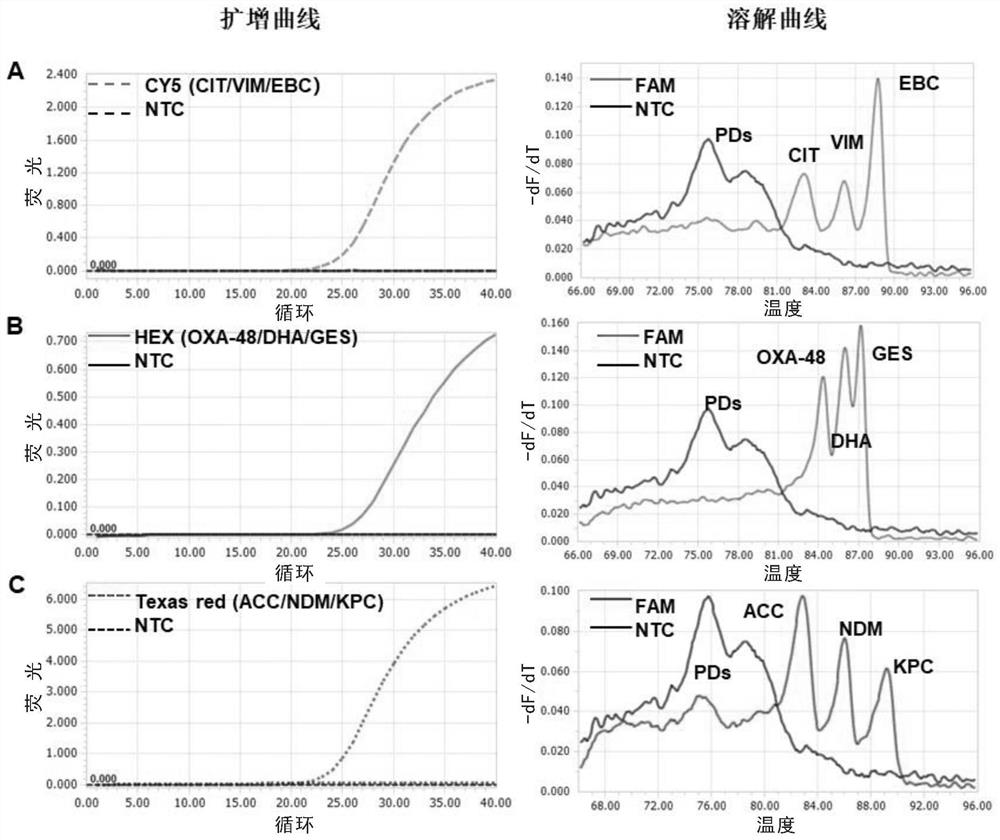
[0142] Example 2. Detection of three mixed templates of Texas red, CY5 and HEX channels by single-tube multiplex fluorescence qPCR method
[0143] In this example, the plasmids of the synthesized genes KPC (SEQ ID NO: 1), NDM (SEQ ID NO: 2) and ACC (SEQ ID NO: 3) are used to mix at 1:1:1 to configure a mixed template Mix1, VIM (SEQ ID NO: 4), EBC (SEQ ID NO: 5) and CIT (SEQ ID NO: 6) plasmids, mixed at 1:1:1 to configure the mixed template Mix 2, OXA48 (SEQ ID NO:7) , GES (SEQ ID NO: 8) and DHA (SEQ ID NO: 9) plasmids, mixed at 1:1:1 and configured into a mixed template Mix 3, using multiplex qPCR to detect the mixed template, primer probes and examples used 1 is the same.
[0144] reaction system:
[0145] 2×PCR buffer (with enzyme) 12.5μl SYTO 9 (10μM) 1 Forward primer (10μM) 0.25*9 Probe (10μM) 0.1*9μl Reverse primer (10μM) 0.25*9μl template DNA10 6
3μl Sterilized distilled water Add to 25μl
[0146] The PCR rea...
Embodiment 3
[0152] Example 3. Sensitivity determination of single-tube multiplex fluorescence qPCR detection
[0153] This example uses the synthesized genes KPC (SEQ ID NO: 1), NDM (SEQ ID NO: 2), ACC (SEQ ID NO: 3), VIM (SEQ ID NO: 4), EBC (SEQ ID NO: 5) ), CIT (SEQ ID NO: 6), OXA48 (SEQ ID NO: 7), GES (SEQ ID NO: 8) and DHA (SEQ ID NO: 9), each of which was serially diluted to 10 1 -10 7 copies / μl template, 9 genes at different concentrations were detected by multiplex qPCR, and the primers and probes used were the same as those in Example 1.
[0154] reaction system:
[0155] 2×PCR buffer (with enzyme) 12.5μl SYTO 9 (10μM) 1 Forward primer (10μM) 0.25*9 Probe (10μM) 0.1*9μl Reverse primer (10μM) 0.25*9μl template DNA10 6
3μl Sterilized distilled water Add to 25μl
[0156] The PCR reaction program is as follows:
[0157] 95°C for 15 minutes; 40 cycles: 95°C for 3 seconds, 60°C for 20 seconds (to collect fluorescence signal...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com