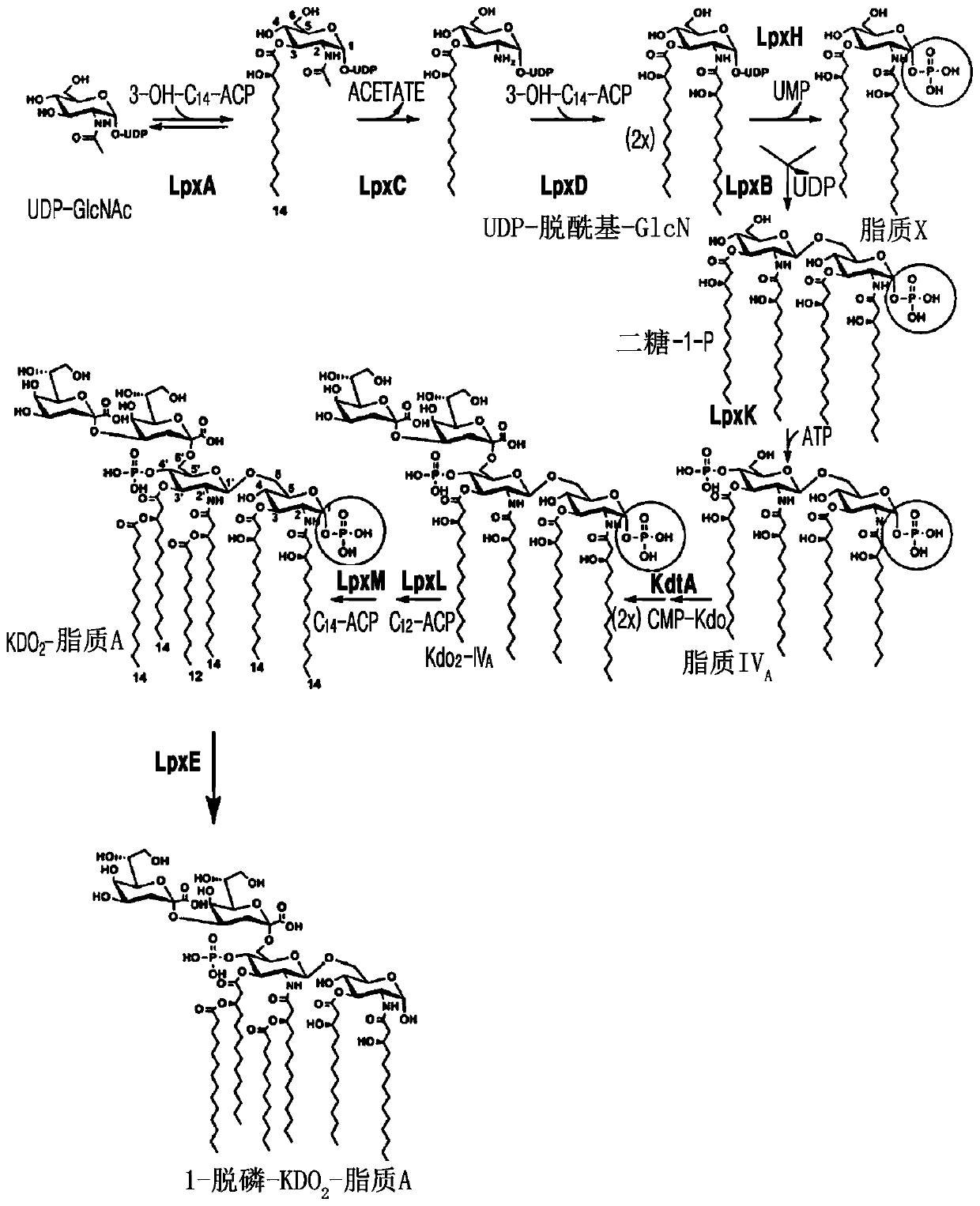
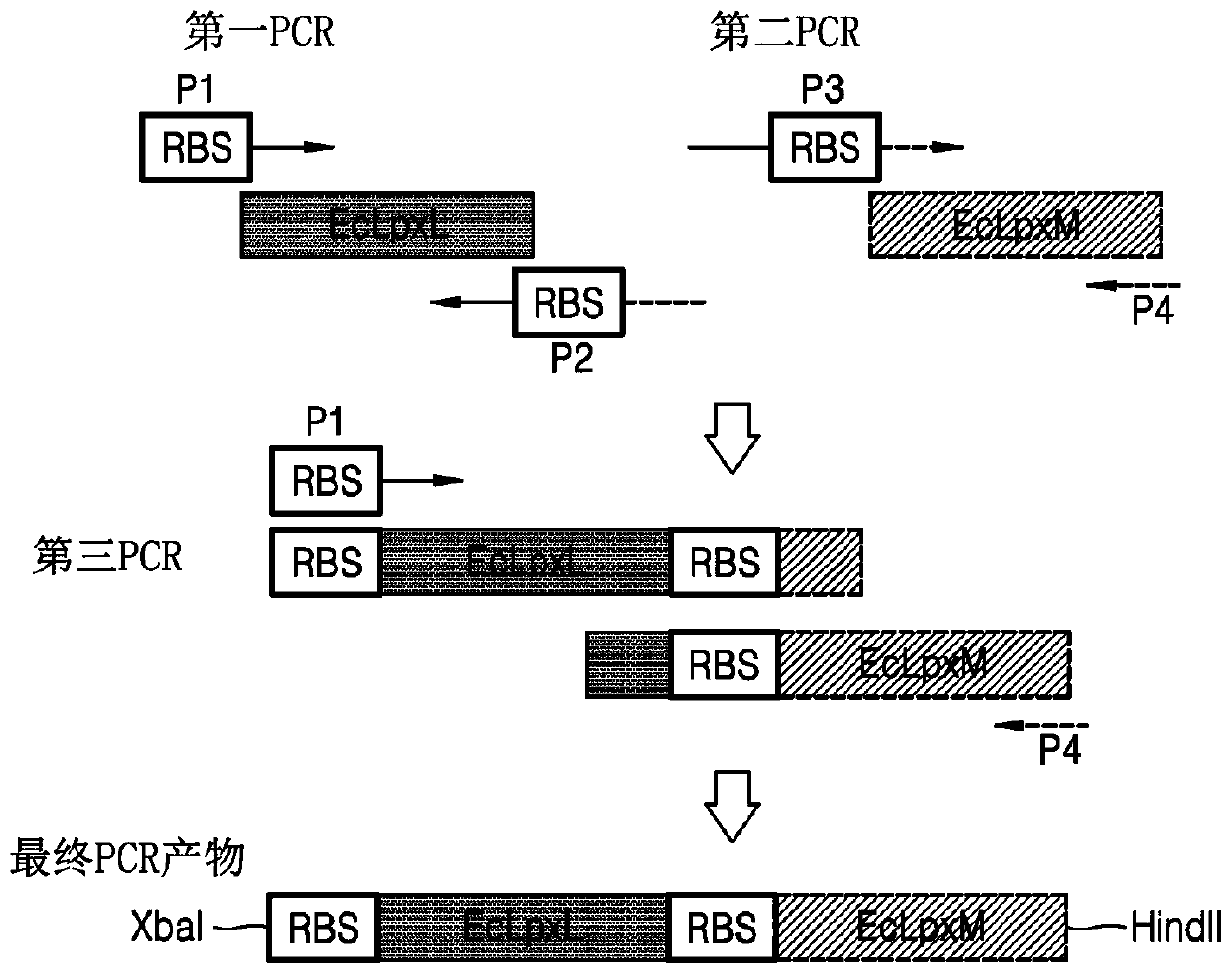
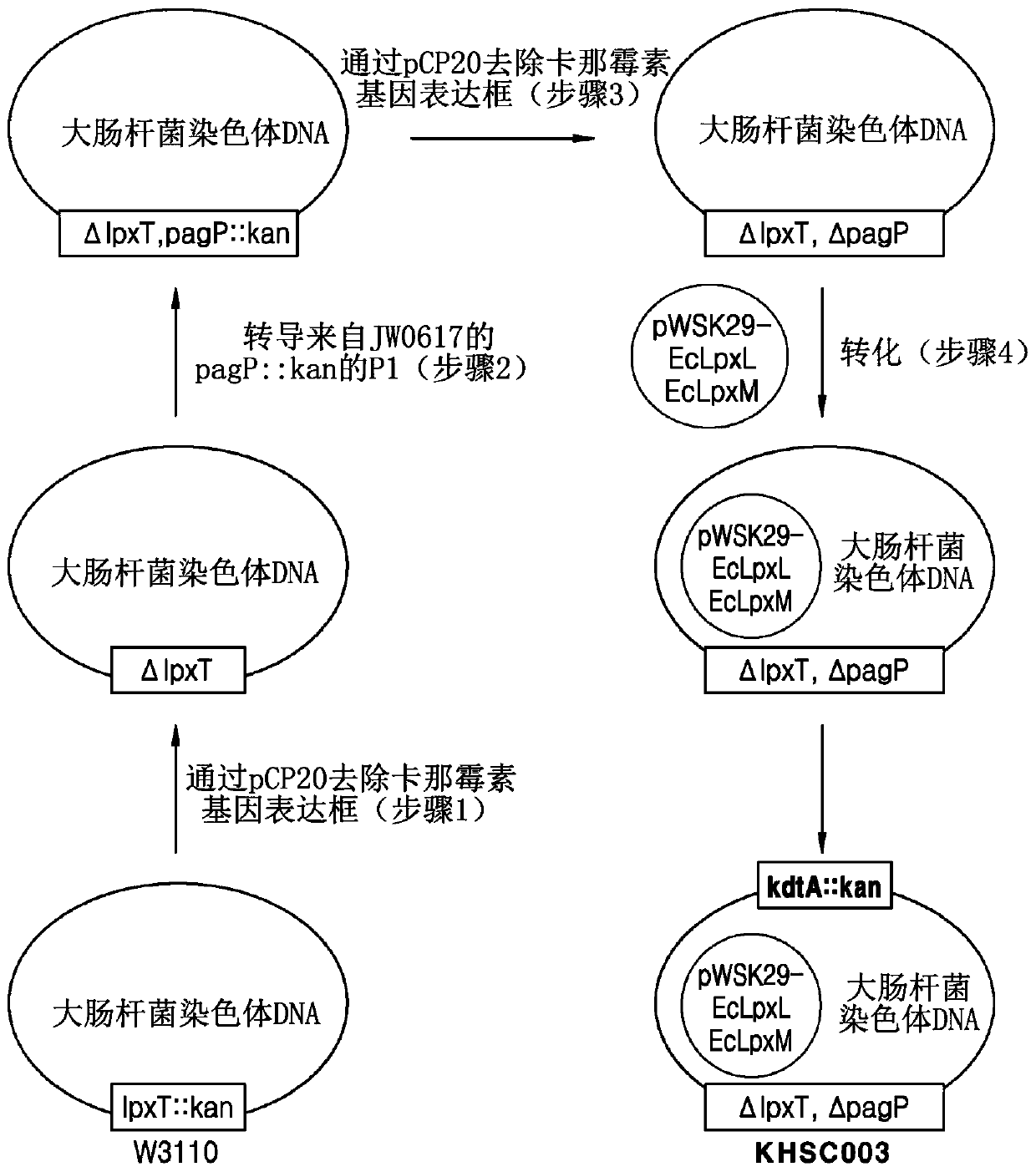
Bacterium producing monophosphoryl lipid A and method of producing monophosphoryl lipid A by using bacterium
A monophosphoryl lipid and bacterial cell technology, applied in biochemical equipment and methods, DNA/RNA fragments, recombinant DNA technology, etc., can solve problems such as low yield and complicated process steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0051] Example 1, preparation of vectors comprising polynucleotides encoding Escherichia coli LpxL and Escherichia coli LpxM
[0052] 1.1. Preparation of pWSK29-EcLpxLEcLpxM
[0053]In order to obtain the polynucleotide encoding the E. coli LpxL polypeptide from the E. coli W3110 genome (GenBank accession number NC_000918.1, ATCC), the encoding was amplified by first polymerase chain reaction (PCR) using a pair of primers shown below. Polynucleotide (GenBank Accession No. AP009048.1 (c1118159.1117239, SEQ ID NO: 2) (see Figure 2A ).
[0054] LpxL forward primer P1: SEQ ID NO: 3
[0055] LpxL reverse primer P2: SEQ ID NO: 4
[0056] In order to obtain polynucleotides encoding E. coli LpxM polypeptides from the E. coli W3110 genome, a pair of primers shown below were used to amplify the EcLpxM polypeptides encoding EcLpxM including RBS (GenBank accession number BAA15663.1, SEQ ID NO: 5) polynucleotide (GenBank accession number AP009048.1 (c1941907.1940936, SEQ ID NO: 6) (se...
example 2
[0080] Example 2, Preparation of Escherichia coli KHSC003 (pWSK29-EcLpxLEcLpxM, kdtA::kan, ΔlpxT, ΔpagP, W3110) strain
[0081] 2.1. Preparation of Escherichia coli with the lpxT gene removed from the genome
[0082] As described in 1.1, the pCP20 plasmid (Kirill A. Datsenko and Barry L. Wanner PNAS (2000), Vol. 97, pp. 6640-6645) was transformed into the kanamycin gene expression cassette (kanamycin cassette) inserted into the coding The lpxT gene (SEQ ID NO: 21) in the E. coli genome of the LpxT polypeptide (SEQ ID NO: 20) was selected on LB-ampicillin solid medium in E. coli strain W3110, lpxT::kan . The selected Escherichia coli was inoculated on LB solid medium, and it was selected at a temperature of 42°C, thereby preparing an Escherichia coli strain from which expression cassettes of lpxT and kanamycin genes were removed, namely, ΔlpxT, W3110 ( Figure 2B Step 1) in .
[0083] 2.2. Preparation of Escherichia coli with pagP and lpxT genes removed from the genome
[...
example 3
[0090] Example 3. Test of lipids of Escherichia coli introduced with AaLpxE, HpLpxE or FnLpxE
[0091] 3.1. Lipid extraction from Escherichia coli W3110 transformed with pWSK29-FnLpxE by acid hydrolysis
[0092] For comparative experiments, Escherichia coli strain W3110 transformed with pWSK29-FnLpxE and Escherichia coli strain W3110 not transformed with pWSK29-FnLpxE were prepared.
[0093] Specifically, pWSK29-FnLpxE was prepared (Wang, X., Karbarz, M.J., McGrath, S.C., Cotter, R.J. and Raetz, C.R., J Biol Chem (2004), Vol. 279(47), pp. 49470-49478), It amplifies the polynucleotide (gb|CP000439.1|:414941-415660 Francisella novicidaU112, SEQ ID NO: 24) encoding FnLpxE polypeptide (gi|118422929|gb|ABK89319.1, Francisella novicida U112, SEQ ID NO: 24) ID NO: 25). The prepared pWSK29-FnLpxE was transformed into Escherichia coli strain W3110 by electroporation, and then the transformed Escherichia coli was selected on LB solid medium containing 50 μg / mL of ampicillin. The sele...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com