Application of CDC20 co-expression gene network as glioma treatment target
A CDC20-M and glioma technology, applied in the biological field, can solve problems such as genome instability, gene site mutation, and chromosome instability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
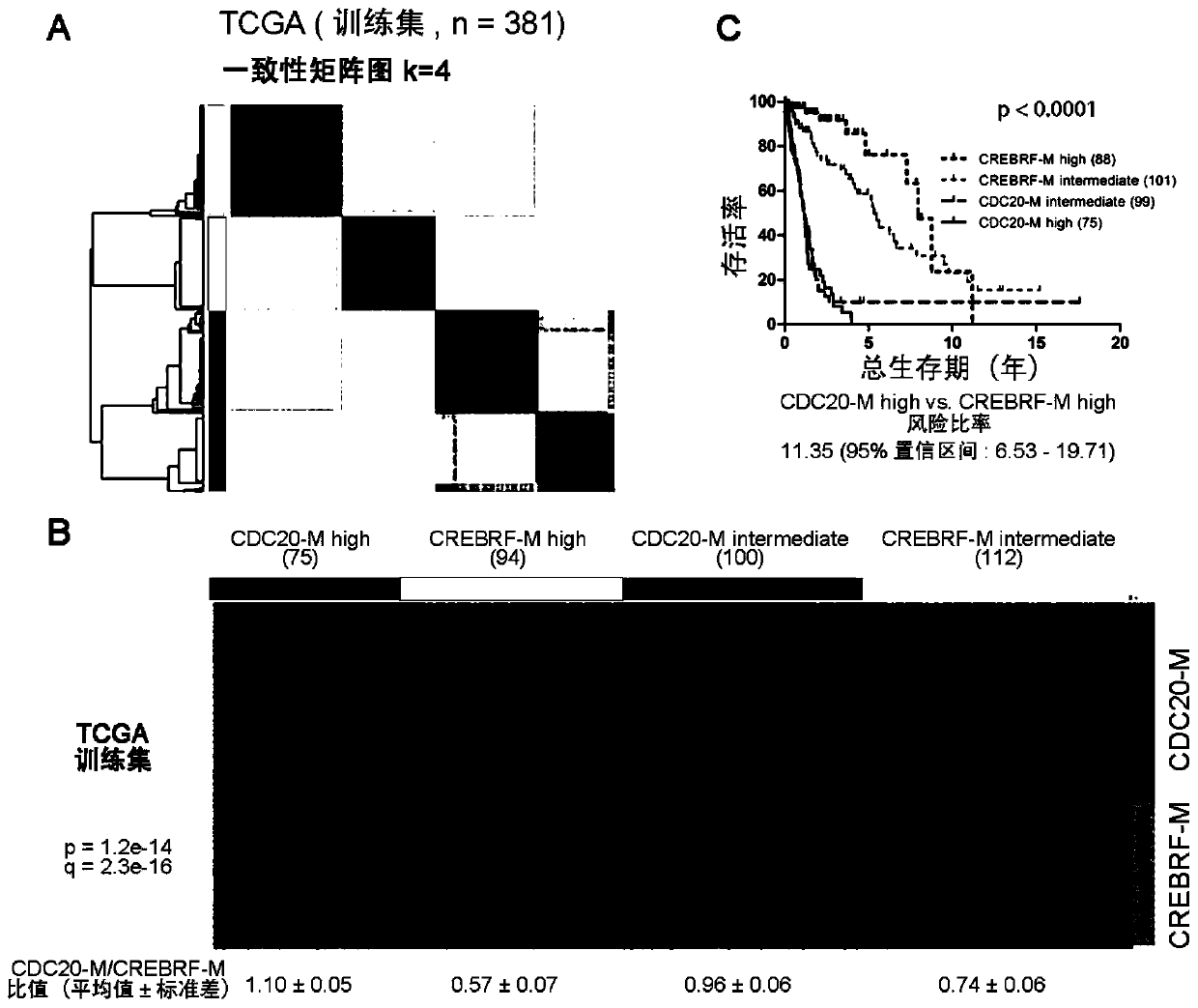
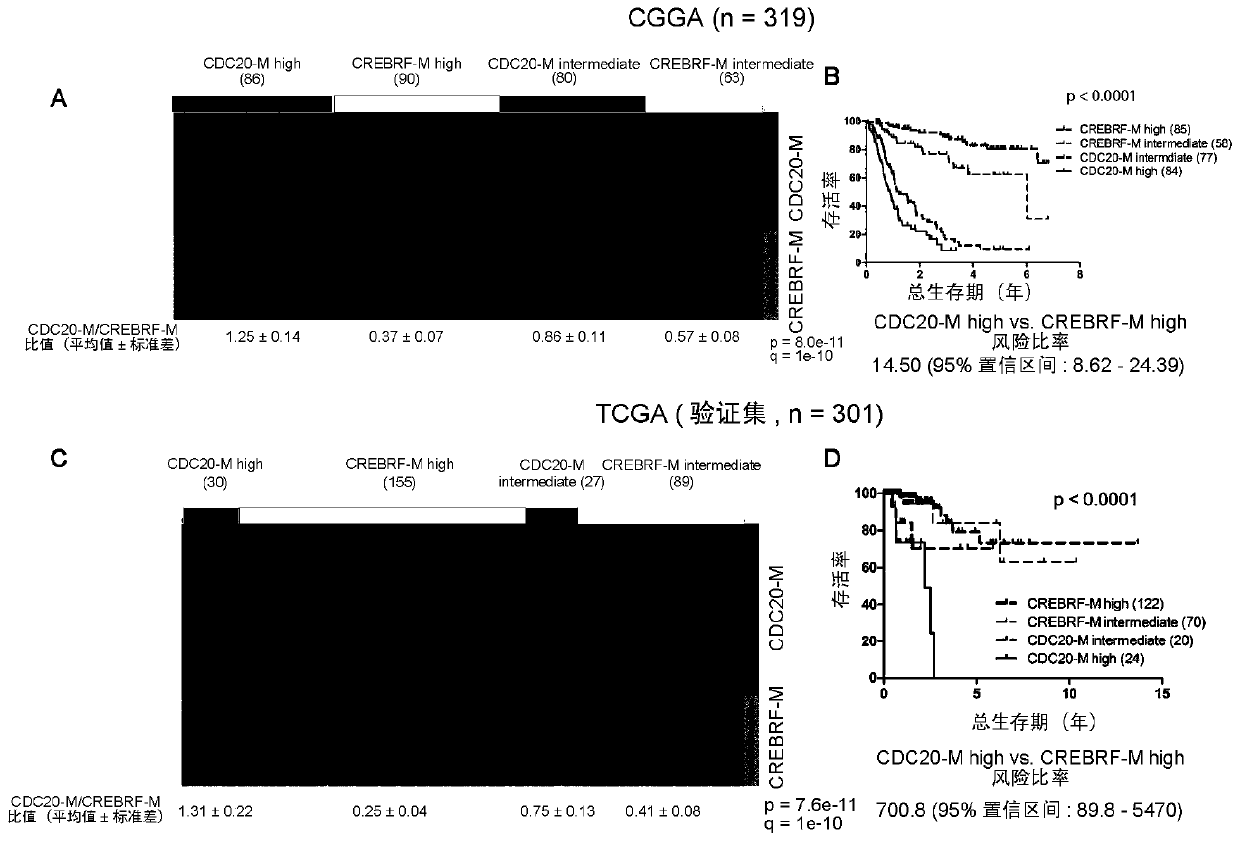
[0074] Example 1, CDC20-M is a sensitive marker of poor prognosis in glioma
[0075] 1. Establishment of CDC20-M and CREBRF-M gene co-expression modules
[0076] The genomic instability of tumors is manifested in the fact that tumor samples carry a large number of dynamically changing chromosomal abnormalities and gene mutations. Different tumor samples may carry different chromosomal abnormalities and gene mutations, and tumors may accumulate important genomic abnormalities at a certain developmental point, and then their genomes remain relatively stable. Therefore, how to identify tumor samples with unstable genomes has become a key issue in this field. One possible strategy is to screen gene expression profiles that correlate with the number of chromosomal variants in representative samples.
[0077] In order to screen specific gene groups that are closely correlated with genome instability in glioma, the present invention uses Pearson correlation analysis, and takes Pear...
Embodiment 2
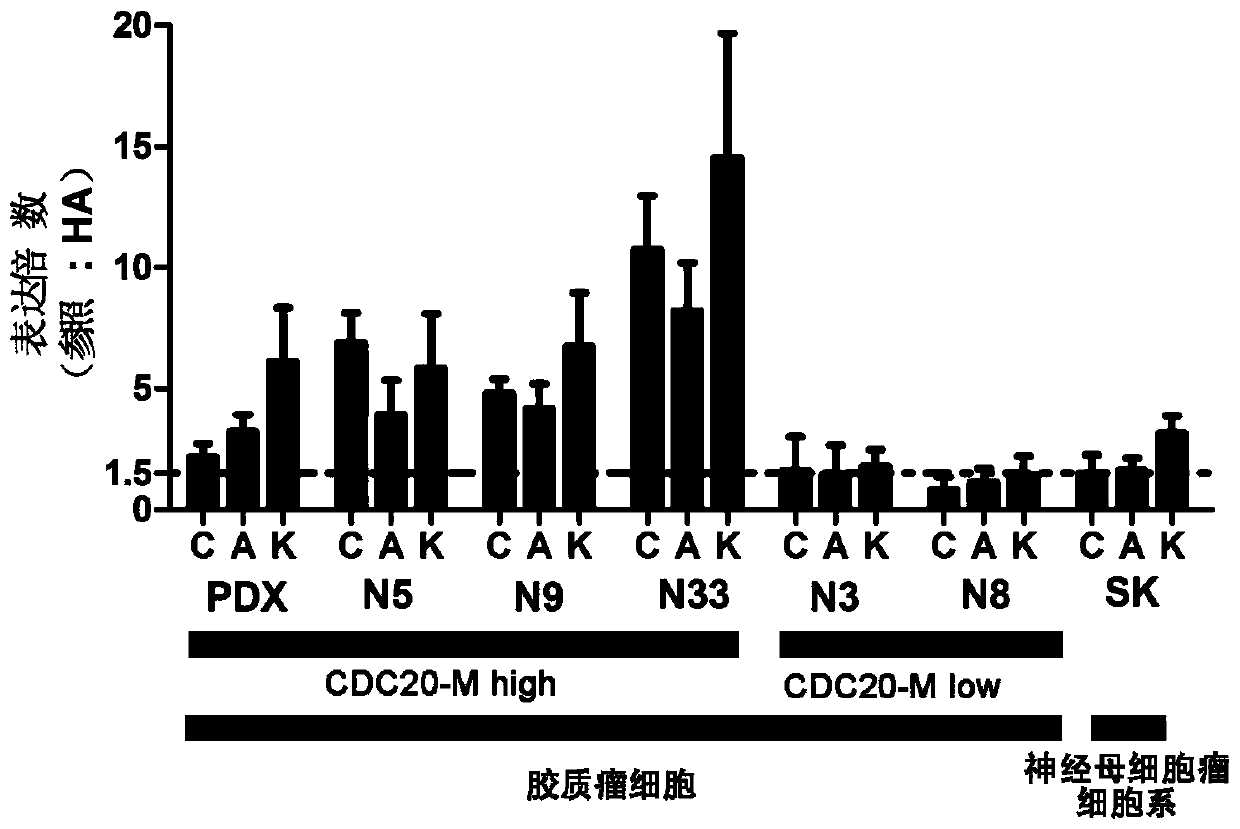
[0099] Example 2, CDC20 co-expression gene network as a therapeutic target for glioma
[0100] 1. CDC20-M typing and identification of glioma cells
[0101] The present invention verifies CDC20-M at the level of cytology. The cells used include: 5 glioma cells (N5, N9, N33, N3 and N8) subcultured after primary culture of glioma blocks resected by surgery; 1 PDX-derived glioma cell (PDX cell ), the genome identification of the above six glioma cells were all IDH1 / 2 wild-type cells; a neuroblastoma cell line SK-N-SH (SK) was used as a control for other tumors, and human as a normal reference Astrocyte Human astrocytes (HA).
[0102] All the glioma cells used, except the PDX cells, the other five cells (N5, N9, N33, N3 and N8) were obtained from the patient's glioma tissue block from Beijing Tiantan Hospital (the cooperative unit has completed the ethical demonstration) , obtained through primary culture and subculture in our laboratory. PDX cells were obtained from patients ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com