A method for analyzing single-cell immune repertoire sequencing data
A technology of immune repertoire and analysis method, which is applied in the field of analysis of single-cell immune repertoire sequencing data, can solve problems such as sequencing errors, insufficient sequencing depth, and insufficient sequencing length, and achieve precise immune function, rich content, and development promotion. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
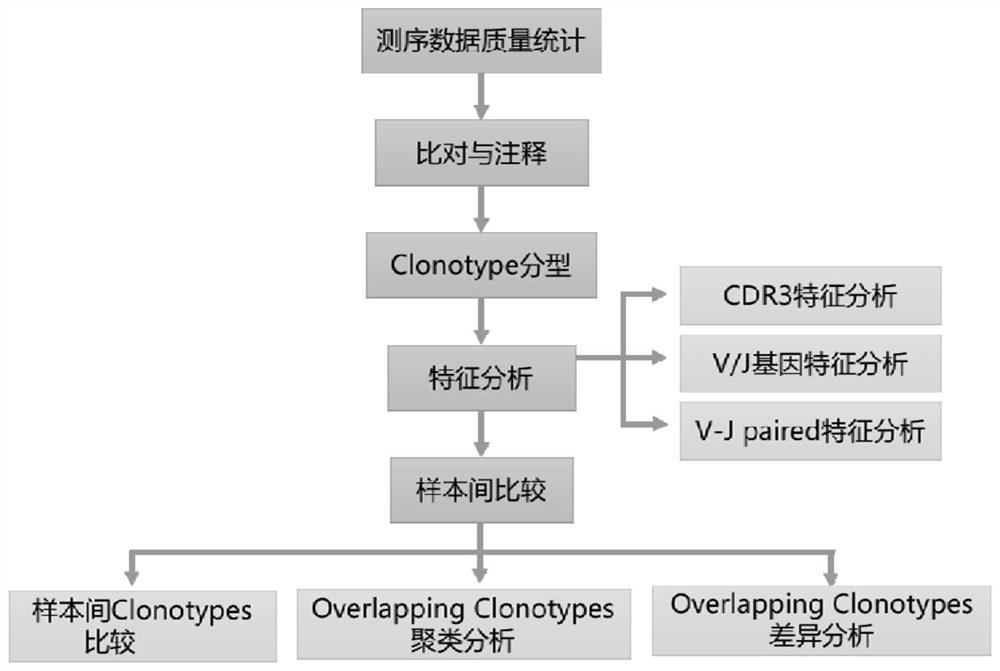
[0051] An embodiment of the method for analyzing single-cell immune repertoire sequencing data of the present invention is a 10X single-cell immune repertoire sequencing data analysis process, figure 1 It is a flow chart for 10X single-cell immune sequencing data analysis, including the following steps:
[0052] Step S1 , perform data quality statistics and basic quality control on the raw data (the original image data generated by the sequencer is converted into sequence data by base calling, including the sequence of the reads and the sequencing quality of the reads): use the official analysis software Cell Ranger of 10XGenomics to analyze the raw data Data quality statistics and basic control to remove reads containing sequencing adapters, unknown bases and low quality, screen out high-quality UMI and high-quality Barcode, and obtain high-quality reads;
[0053] Step S2 , data comparison and annotation, including steps:
[0054] S2.1 Data comparison: Cell Ranger first ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com