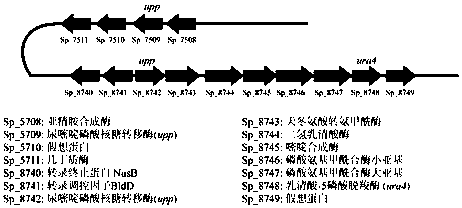
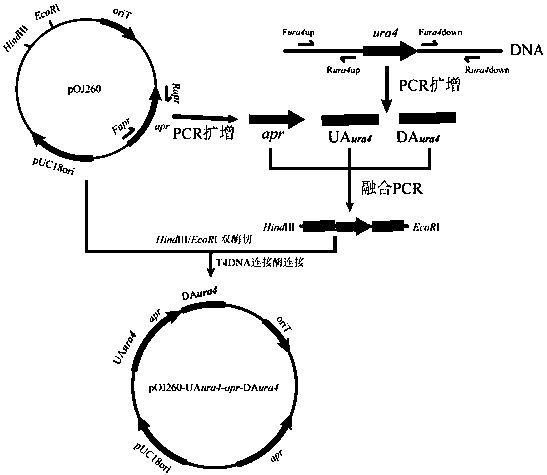
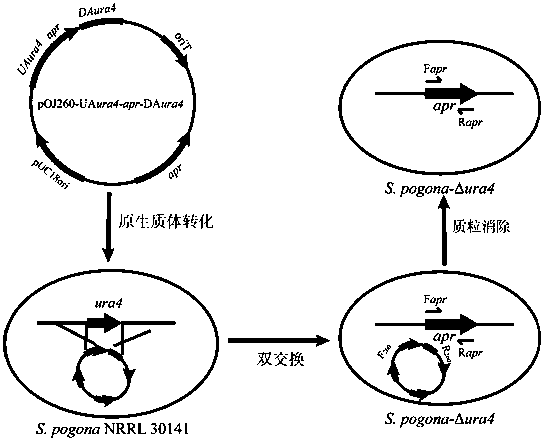
Method of blocking gene clusters of Saccharopolyspora pogona based on homologous recombination of linear fragments
A homologous recombination technology of Saccharopolyspora spp., applied in the field of gene cluster blocking of S. spp., can solve problems such as retention and difficulty in exogenous plasmid transformation, and achieve the effect of increasing production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] (1) Specific medium formula and culture conditions
[0042] (1) S. pogona seed activation medium CSM (Complete synthetic medium): TrypticSoyBroth4.5g / L, Glucose 0.9g / L, Yeast extract 0.3g / L, MgSO 4 .7H 2 O 0.22g / L. Inoculate the monoclonal or 2% strain preservation solution into a 150 mL shake flask equipped with 20 mL LCSM, and incubate at 30°C and 280r / min for 48 h;
[0043] (2) S. pogona synthetic fermentation medium (Synthetic fermentation medium, SFM): Glucose20.0 g / L, Yeast extract 4.0 g / L, Tryptone 4.0 g / L, KNO 3 1.0 g / L, K 2 HPO 4 •3H 2 O 0.5g / L, MgSO 4 •7H 2 O 0.5 g / L, FeSO 4 •7H 2 O 0.01 g / L. Inoculate 2% strain activation solution into a 300mL shake flask filled with 50mL SFM, and culture at 30°C and 280r / min for 12 days.
[0044] (3) R5 medium was used for protoplast transformation: sucrose 103 g / L, glucose 10 g / L, casamino acids 0.1 g / L, Yeast extract 5 g / L, K 2 SO 4 0.25 g / L, MgCl 2 •6H 2 O 10.12g / L, TES 5.73 g / L, trace element solution 2 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com