MGMT gene promoter methylation detecting method, sequencing data processing method and processing device
A technology for sequencing data and processing methods, applied in biochemical equipment and methods, genomics, and microbial determination/inspection, etc., can solve problems such as low detection accuracy, and achieve the effect of facilitating evaluation and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
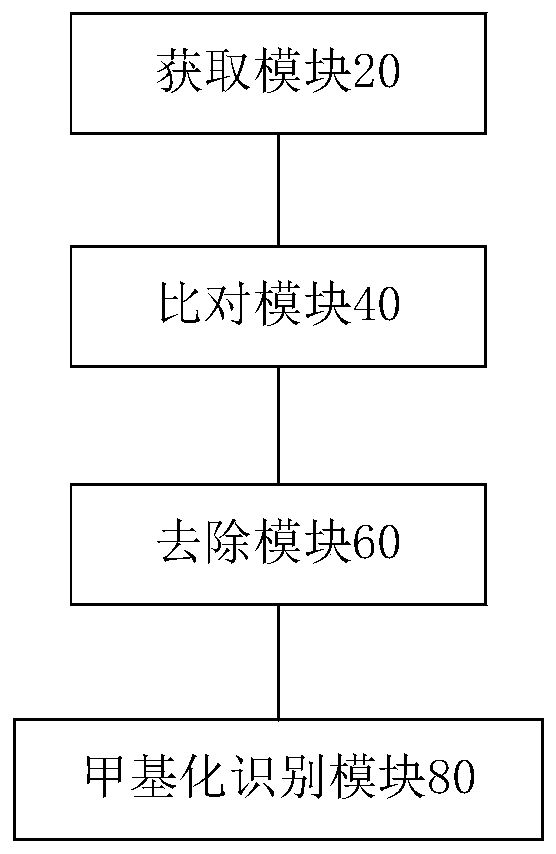
Image
Examples
Embodiment 1
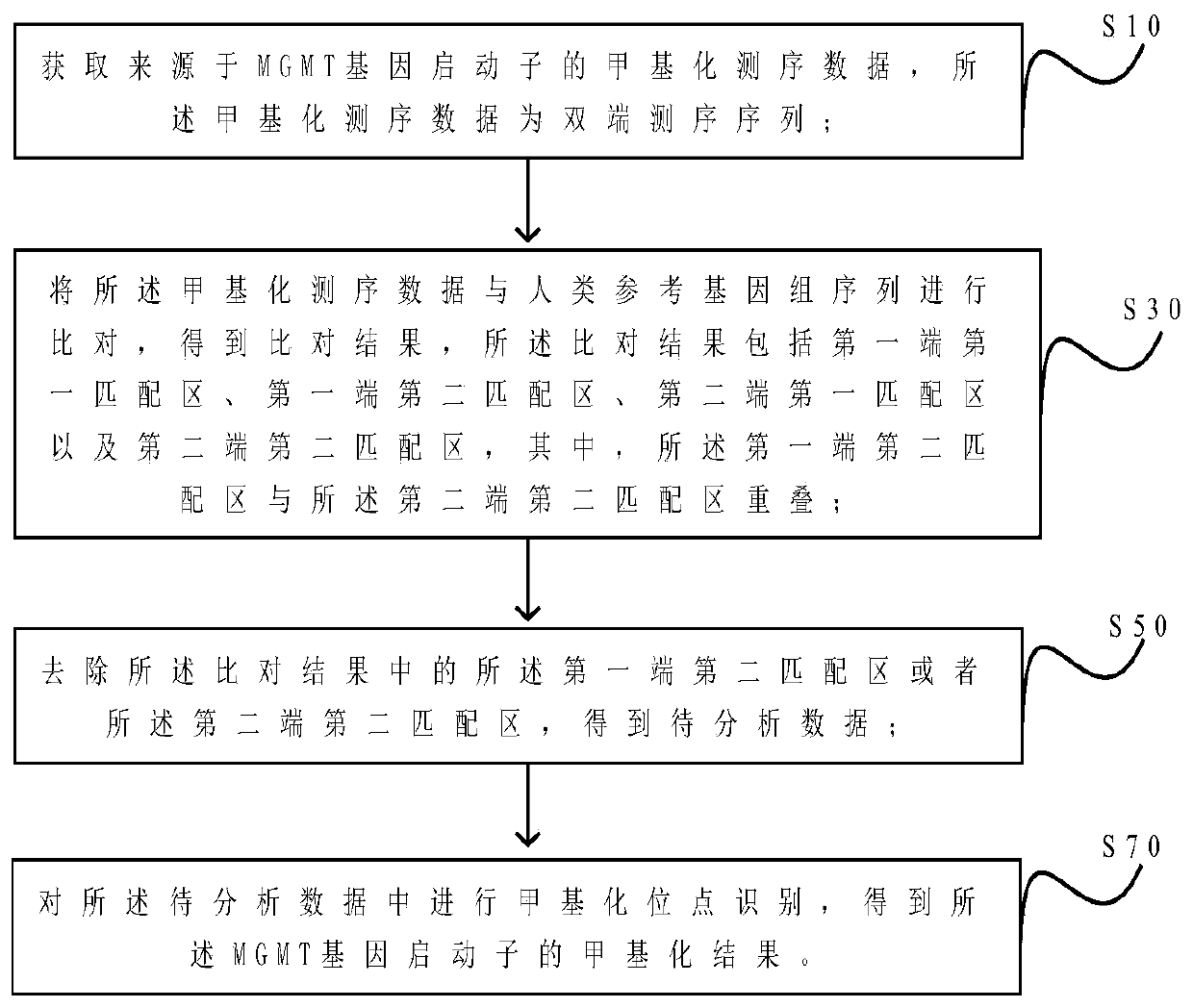
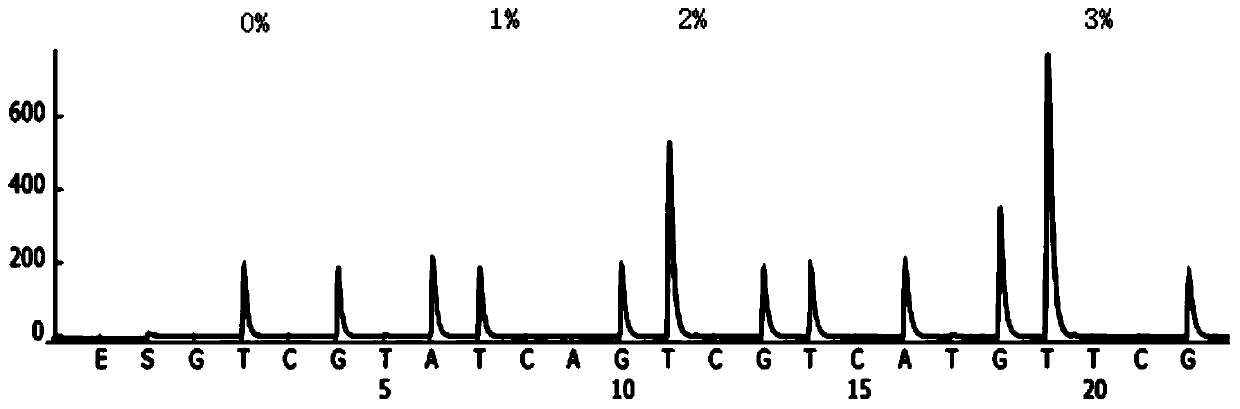
[0072] Example 1: Detection of MGMT gene promoter methylation level
[0073] The primer sequences used in this example are shown in Table 2 below, synthesized by Jinweizhi Company, and diluted with dewatered to a working concentration of 5-10 μM.
[0074] Table 2:
[0075] name SEQ ID NO: Sequence (5'-3') upstream sequence F 1 TYGYGTTTTGGATATGTTGG downstream sequence R 2 CRAAAAAAAACTCCRCACTC
[0076] Specific steps are as follows:
[0077] 1. Extract the genomic DNA of the sample to be tested.
[0078] 2. Bisulfite transforms genomic DNA.
[0079] 2.1 The initial amount of transformed DNA is 100ng, and the initial volume of the sample is 20μL. If it is less than 20μL, make up with water.
[0080] 2.2 Take 130 μL Lightning Conversion Reagent and add it to the DNA sample, vortex to mix, centrifuge briefly, put it on the PCR machine, and perform the PCR reaction as shown in Table 3:
[0081] table 3:
[0082] temperature time ...
Embodiment 2
[0102] Embodiment 2: Detect the amplification effect of the annealing temperature, working concentration and PCR cycle number of MGMT gene promoter methylation primer
[0103] 1. Extraction of tissue DNA from clinical samples.
[0104] 2. Refer to Example 1 for steps such as bisulfite conversion of genomic DNA and MGMT amplification.
[0105] 3. Choose different primer annealing temperature, working concentration and number of PCR cycles.
[0106] 3.1 Choice of primer annealing temperature: 40°C, 45°C, 50°C, 55°C, 60°C.
[0107] 3.2 Choice of primer working concentration: 4 μM, 5 μM, 10 μM, 15 μM, 16 μM.
[0108] 3.3 Selection of the number of PCR cycles: 25 cycles, 30 cycles, 35 cycles, 40 cycles, 45 cycles.
[0109] 4. Test results:
[0110] 4.1 The detection results of primer annealing temperature are shown in Table 6:
[0111] Table 6:
[0112] Annealing temperature Test results 40℃ More non-specific amplification 45℃ Amplify the correct targe...
Embodiment 3
[0120] Example 3: Processing method of MGMT gene methylation sequencing data
[0121] 1. Comparison
[0122] Call bismark to compare each pair of fastq files to the MGMT human reference genome sequence as paired reads, generate an initial bam file, and set the parameter "--phred33-quals".
[0123] 2. Sorting
[0124] Call the sort module of SAM tools to sort the initial bam file according to the chromosome position, with default parameters.
[0125] 3. Add Read Group information
[0126] Call the Add Or Replace Read Groups module of Picard to add ReadGroup information to the sorted bam file, and set the parameter "VALIDATION_STRINGENCY=LENIENT".
[0127] 4. Remove the overlapping interval between the paired-end sequences
[0128] Call the clip Overlap module of Bam Util to remove the overlapping sequences between the paired-end sequences in the bam file after alignment. In the subsequent analysis, this part of the overlapping sequences will not be filtered, which will affe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com