A multi-gene combined detection reagent
A joint detection and multi-gene technology, applied in the field of biomedicine, can solve the problems of the sensitivity not reaching more than 90%, the overlap of colorectal cancer detection value and performance, and the difficulty of gene methylation markers, so as to achieve easy and reliable acquisition High diagnostic and detection sensitivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0137] According to Table 1, three pairs of primers for SDC2 were designed to study the specificity of amplification.
[0138] Table 1 Primers for SDC2
[0139]
[0140]
[0141] The above three pairs of primers were used to detect the DNA of stool samples from 2 cases of colorectal cancer patients and 2 cases of normal human stool samples, and analyze the specificity of amplification.
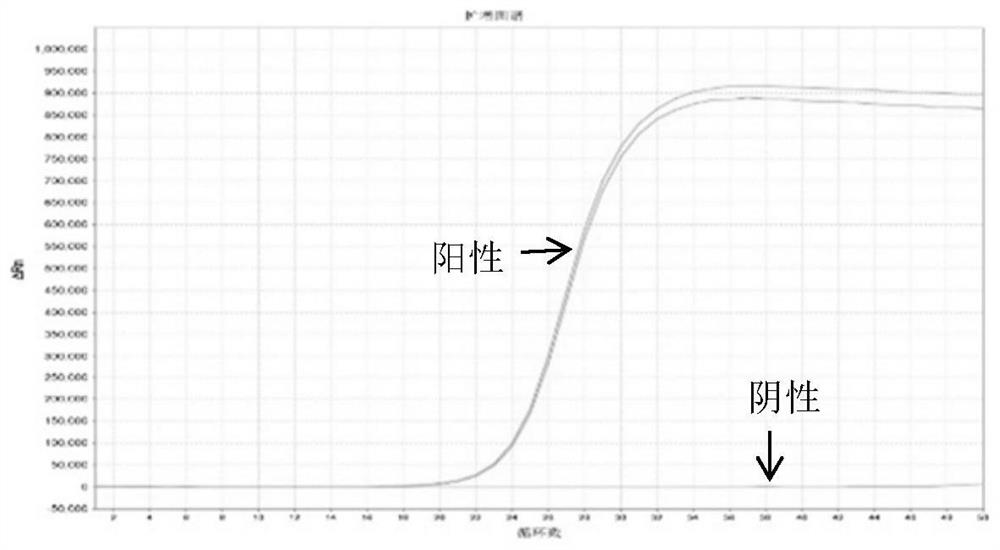
[0142] The amplification curve of 3 pairs of primers amplifying SDC2 is as follows Figure 1-3 shown.
[0143] The results showed that SDC2 primers had a better amplification specificity for S0; while the other two pairs of primers had little difference between negative and positive samples in their amplification curves, and could not effectively distinguish between negative and positive samples.
Embodiment 2
[0145] According to Table 2, three pairs of COL4A1 / COL4A2 primers were designed to study the specificity of amplification.
[0146] Table 2 Primers for COL4A1 / COL4A2
[0147]
[0148] The above 3 pairs of primers were used to detect the DNA of stool samples from 3 cases of colorectal cancer patients and 3 cases of normal human stool samples, and analyze the specificity of amplification.
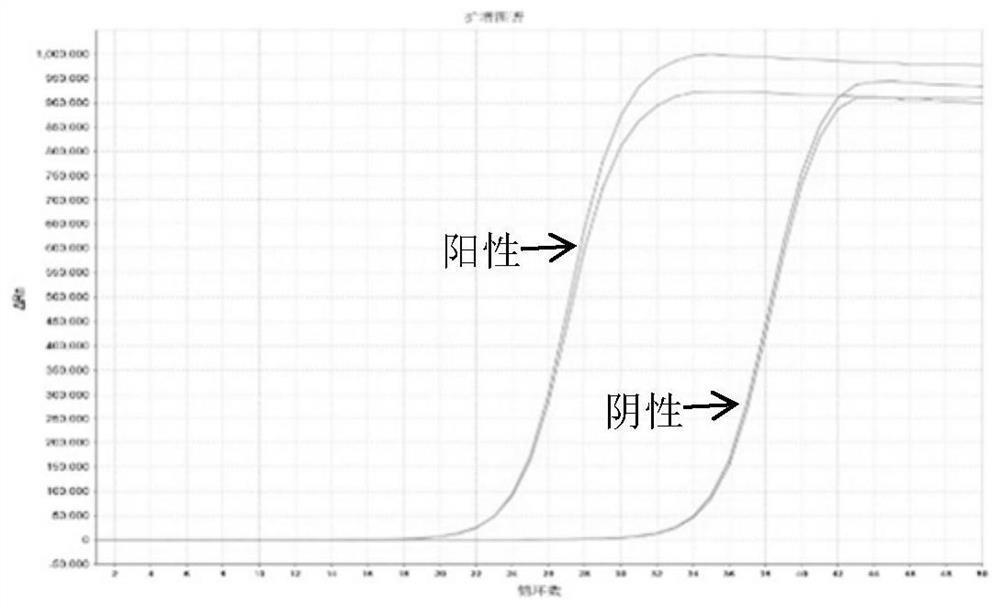
[0149] The amplification curves of 3 pairs of primers amplifying COL4A1 / COL4A2 are as follows: Figure 4-6 shown.
[0150] The results showed that COL4A1 / COL4A2 primer pair C0 and C1 had better amplification specificity; while primer pair C2 had no amplification for COL4A1 / COL4A2.
Embodiment 3
[0152] According to Table 3, three pairs of primers for ITGA4 were designed to study the specificity of amplification.
[0153] Table 3 Primers of ITGA4
[0154]
[0155]
[0156] The above 3 pairs of primers were used to detect the DNA of stool samples from 3 cases of colorectal cancer patients and 3 cases of normal human stool samples, and analyze the specificity of amplification.
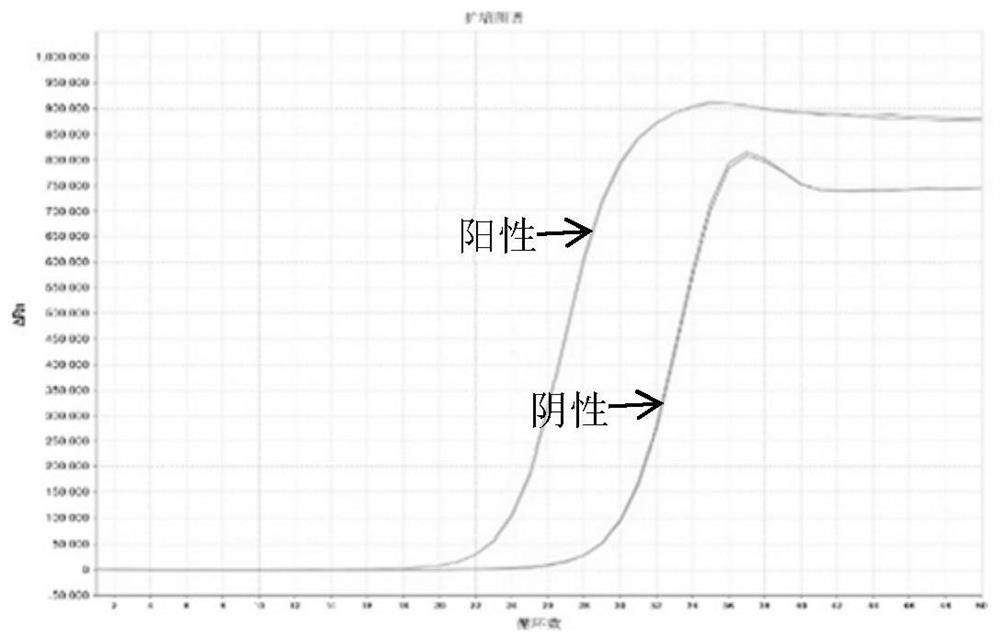
[0157] The amplification curve of 3 pairs of primers to amplify ITGA4 is as follows Figure 7-9 shown.
[0158] The results showed that the primer pair I0 and I2 of ITGA4 had better amplification specificity; while the amplification curve of primer pair I1 had little difference between negative and positive samples, and could not effectively distinguish between negative and positive samples.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com