Bladder cancer maker set and application thereof
A technology of bladder cancer and markers, applied in the field of molecular biology, can solve the problems of low sensitivity, error in results, high cost of patients, etc., and achieve the effect of improving sensitivity, easy operation and high acceptance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Sample Handling (Urine Samples)
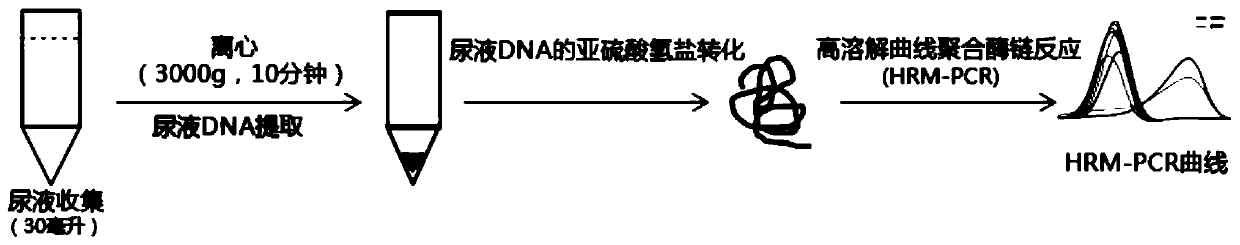
[0038] 1) Collect 30ml of urine in a urine collection tube pre-stored with 600ul EDTA solution, mix well and centrifuge at 3000g for 10 minutes;
[0039] 2) Urine DNA was extracted using a Qiagen kit;
[0040]3) Urine DNA was subjected to bisulfite conversion using the EZ Gold Methylation Kit.
[0041] Urine storage conditions: if the storage time is less than 48 hours, store at 4°C; if the storage time exceeds 48 hours, store at -80°C.
Embodiment 2
[0043] HRM-PCR determination of gene methylation level
[0044] High melting curve polymerase chain reaction (HRM-PCR) experiments were performed as follows:
[0045] 1) Prepare the reagents required for HRM-PCR (1 reaction):
[0046] a.5ul of 2X Zymo premix;
[0047] b. 0.5ul of primers, the primer sequence is shown in SEQ ID NO: 1-14;
[0048] c.0.5ul of LC Green Plus Saturated fluorescent dyes;
[0049] d.3ul of water;
[0050] The sample of e.1ul, this sample has been processed according to the method for embodiment 1;
[0051] 2) Perform HRM-PCR assay to obtain gene methylation level, the reaction steps and conditions are as follows:
[0052] a. Preactivation at 95.0°C for 10 minutes;
[0053] b. Denaturation at 95.0°C for 15 seconds, annealing at 60.0°C for 30 seconds, extension at 72.0°C for 15 seconds, and 50 cycles;
[0054] c. Obtain the HRM curve according to the following reaction conditions: raise the temperature from 72.0°C to 95.0°C at a rate of 1.6°C / ...
Embodiment 3
[0056] Construction of bladder cancer risk score model
[0057] 1. Sample collection
[0058] Eighty-one human urine samples were collected, including 44 bladder cancer patients and 37 normal controls.
[0059] 2. Model Construction
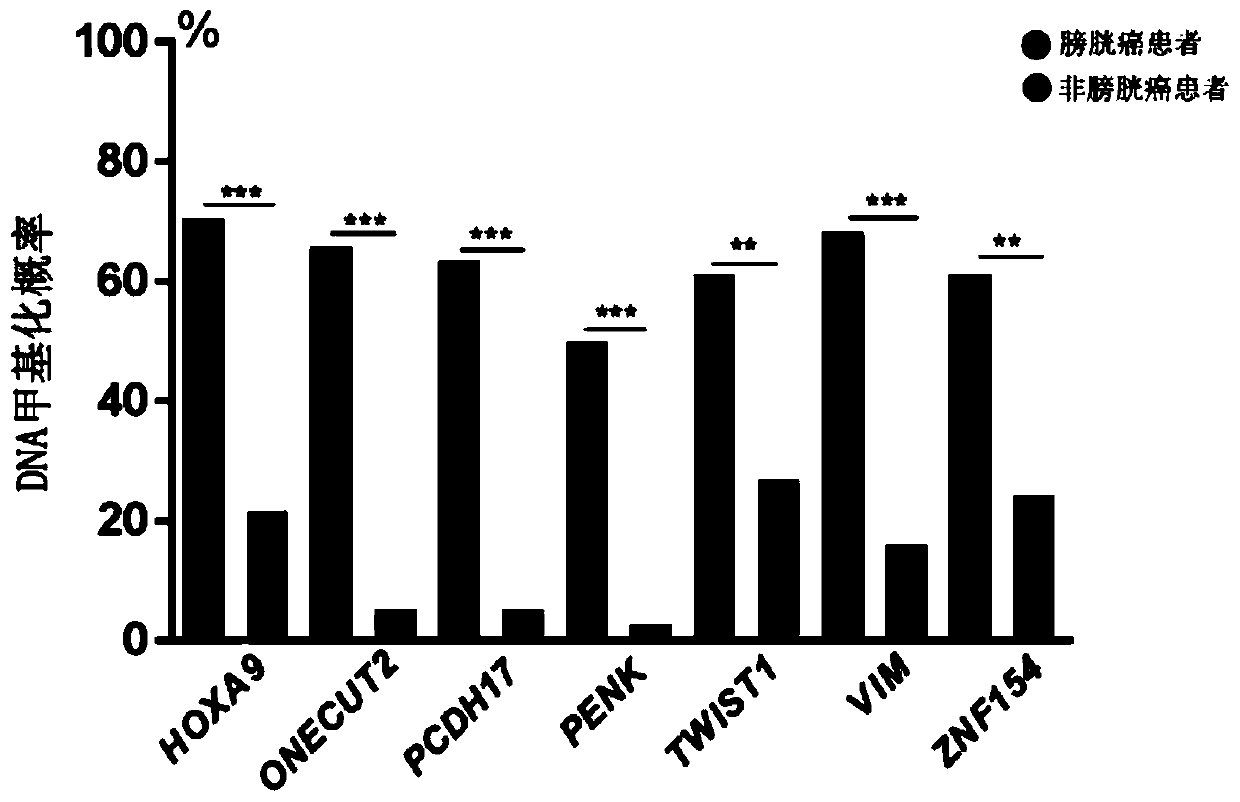
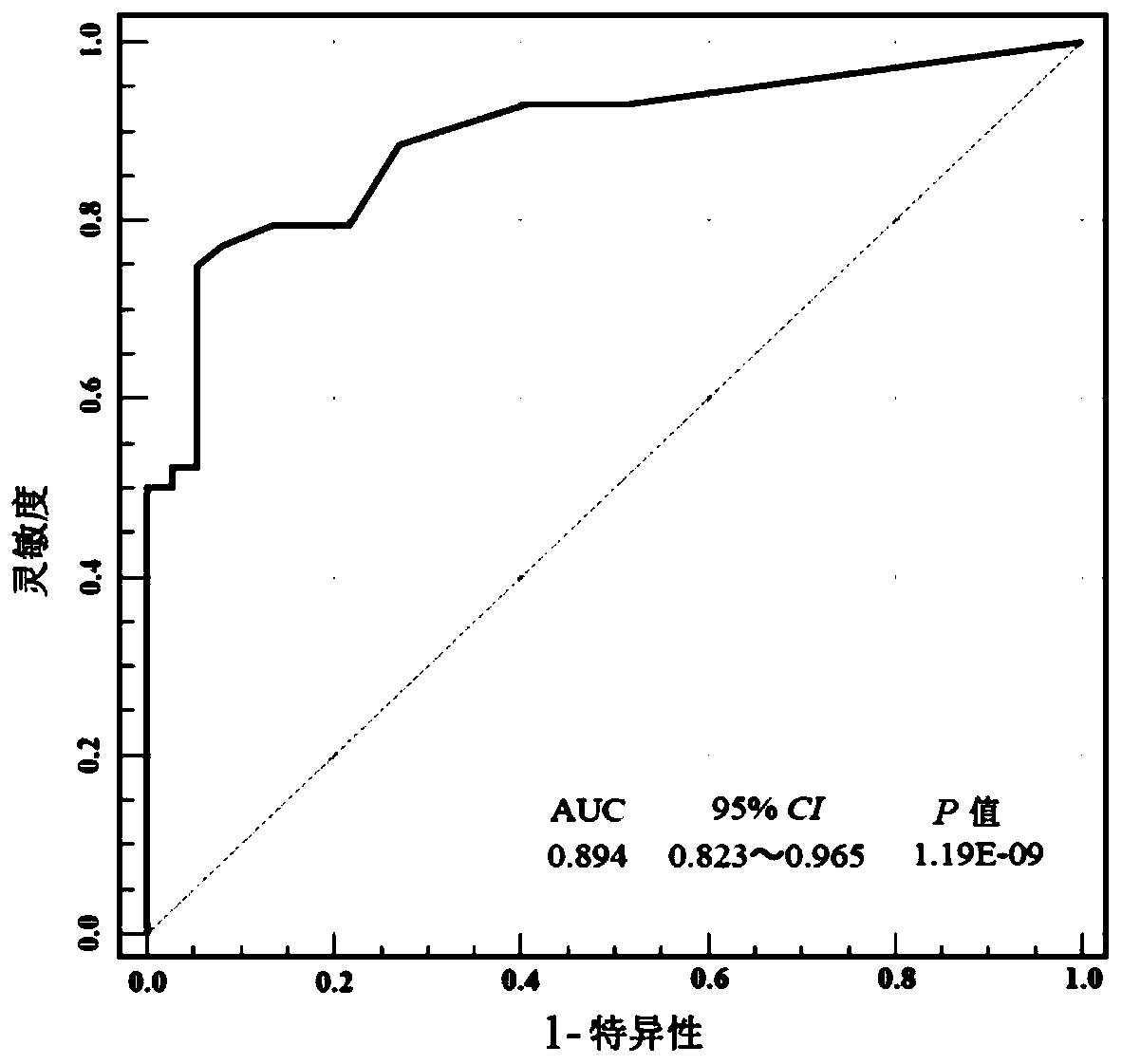
[0060] Univariate and multivariate logistic regression analyzes were performed on the methylation levels of 7 selected genes (HOXA9, ONECUT2, PCDH17, PENK, TWIST1, VIM, and ZNF154), and a detection model including 7 genes was constructed. Using the method of Example 2, the methylation levels of 7 genes in each sample were obtained respectively, and the operation process of the detection experiment was as follows figure 1 shown. First, a univariate logistic regression analysis was performed. Such as figure 2 As shown, the 7 selected genes were all significantly associated with bladder cancer. Multivariate logistic regression analysis was then performed to obtain the regression coefficients of the seven genes.
[0061] All statistical ana...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com