Method for positioning T-DNA insertion site by flag tag of sam file
A technology for inserting sites and tags, which is applied in the field of sam file flag tags to quickly locate T-DNA inserting sites, which can solve problems such as unsuitable for large-scale promotion, cumbersome steps, complex algorithms, etc., to eliminate false positive results and simple operation , low-cost effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0031] In order to make the object, technical solution and advantages of the present invention more clear, the present invention will be further described in detail below in conjunction with the examples. It should be understood that the specific embodiments described here are only used to explain the present invention, not to limit the present invention.
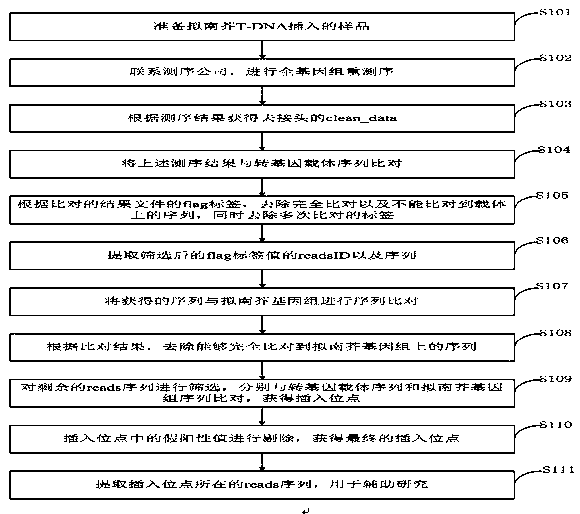
[0032] In order to solve the problem of fast, free and efficient identification of T-DNA insertion sites, the present invention has developed a set of whole genome resequencing technology, using transgenic vectors for sequence comparison, screening according to the Flag tag of the comparison result file, and then A method for comparing the screened sequence with the Arabidopsis genome to finally obtain the T-DNA insertion site. The method has the characteristics of simple operation, time saving and high efficiency, low cost and accurate results.
[0033] The application principle of the present invention will be described ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com