RNA site-directed editing technology based on CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)-Cas13a
An editing and technical technology, applied in the field of RNA fixed-point editing, can solve the problems of easy off-target and low binding force
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
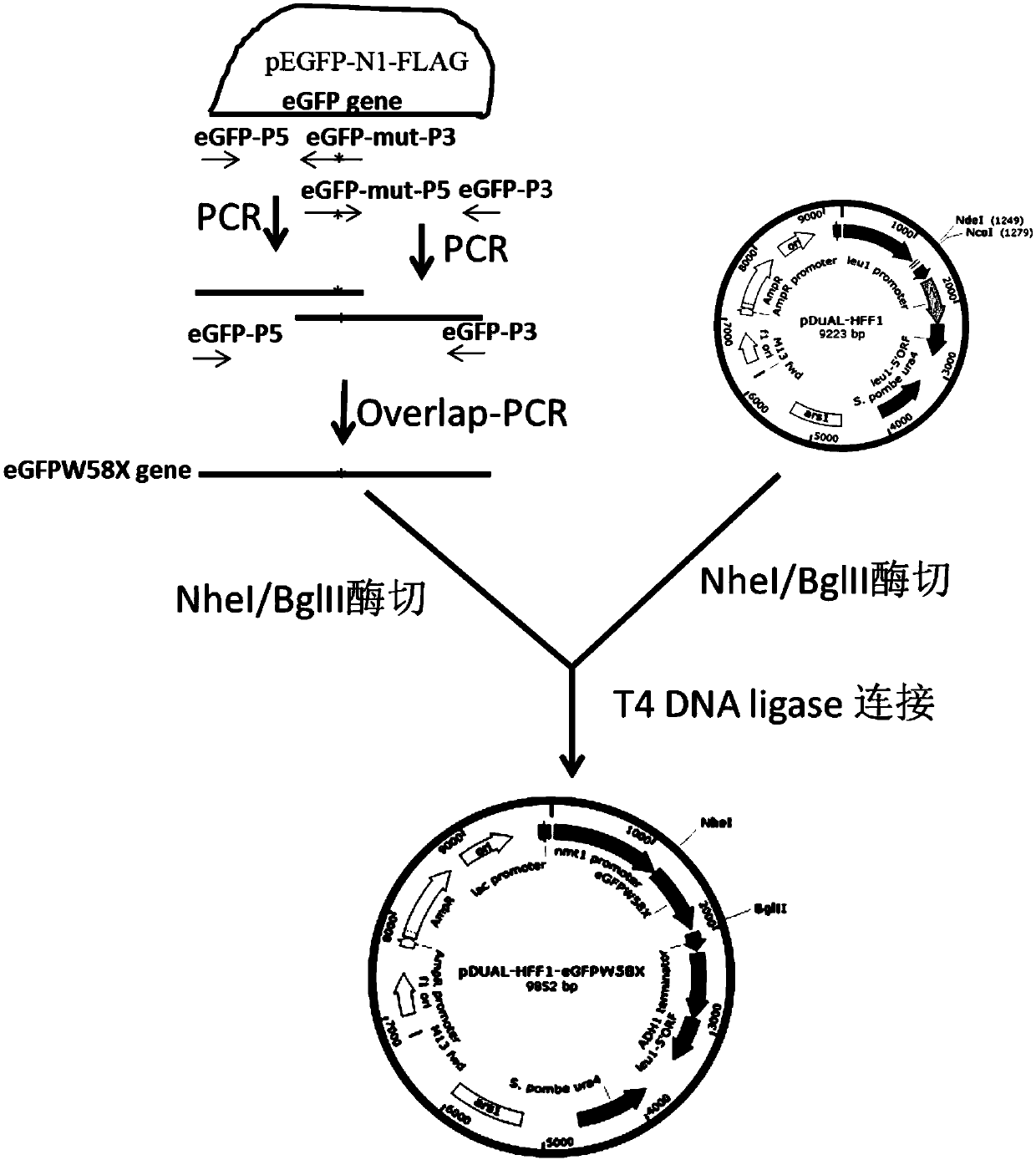
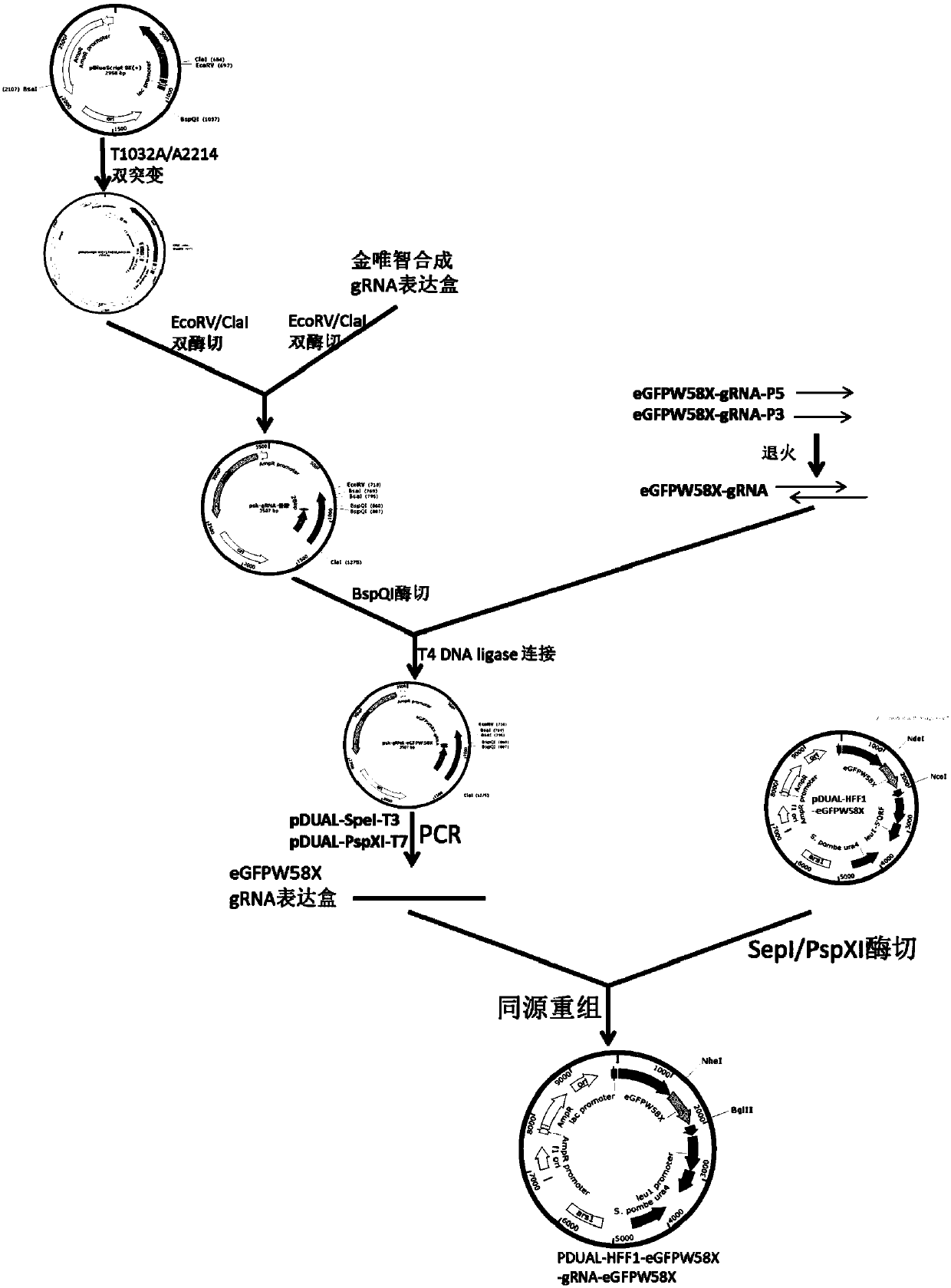
[0068] Example 1. The Crispr-Cas13a-based RNA site-directed editing technology is used to restore the mutated green fluorescent protein gene transcript
[0069]1. Expression of dCas13a-hADAR2d fusion protein in fission yeast cells
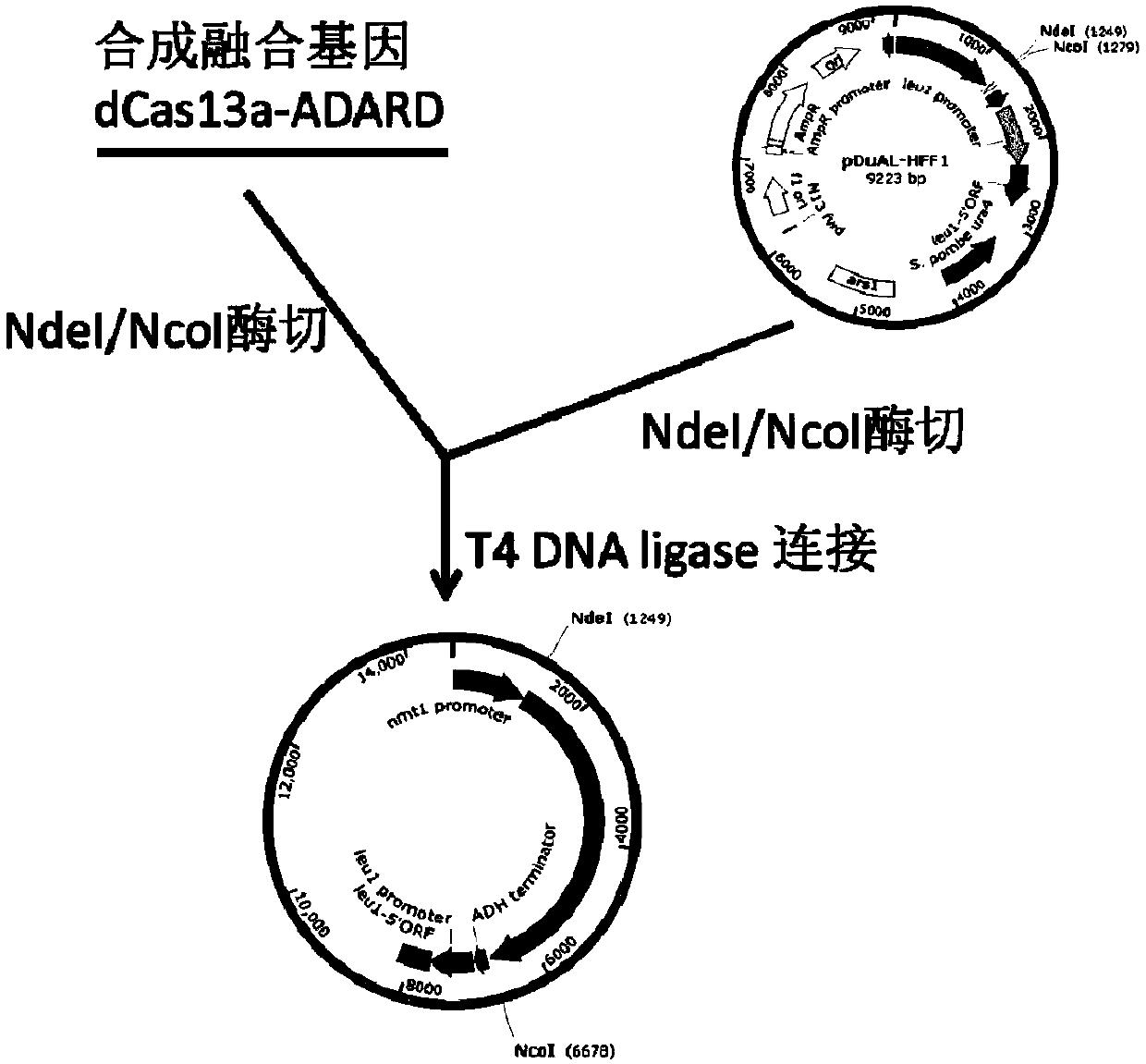
[0070] 1. Construction of pDUAL-HFF1-dCas13a-hADAR2d vector expressing dCas13a-hADAR2d fusion protein
[0071] In the following examples, the Cas13a variant is called "dCas13a", which corresponds to the mutation at position 1278 of its amino acid sequence.
[0072] The gene of dCas13a-hADAR2d fusion protein (dCas13a coding sequence is located at the 5' end) was synthesized by gene synthesis method (Jin Weizhi). The gene sequence includes the yeast linker sequence (underlined part), and the sequence is as follows (SEQ ID NO: 1):
[0073]
[0074]
[0075] In the above sequence, the bases in the box are the bases of the mutant sites, and the underlines are the linker sequences. Among them, the 1-1389th position is the dCas13a sequence, and th...
Embodiment 2
[0127] Example 2, RNA site-directed editing technology based on Crispr-Cas13a is used for fission yeast endogenous gene tdh1 transcript
[0128] 1. Expression of dCas13a-hADAR2d fusion protein in fission yeast cells
[0129] Using the same method as in Example 1, the dCas13a-hADAR2d fusion protein was expressed in fission yeast cells.
[0130] 2. Expression of CRISPR-Cas13a crRNA targeting tdh1 gene transcript in fission yeast cells
[0131] 1. Design primers targeting the crRNA of the tdh1 gene, the sequence is as follows (capital letters indicate crRNA):
[0132] tdh1-crRNA-P5:aacGAATTGCCATTTTGAATCAAGTGTAAATCAATACCATGGATGAATGATCTATACAGAAGCGATGC (SEQ ID NO: 13);
[0133] tdh1-crRNA-P3:gccGCATCGCTTCTGTATAGATCATTCATCCATGGTATTGATTTACACTTGATTCAAAATGGCAATTC (SEQ ID NO: 14);
[0134] 2. Construct the plasmid pSK-crRNA(tdh1), the specific operation method is as follows:
[0135] A. Dissolve tdh1-crRNA-P5 and tdh1-crRNA-P3 primers into 10 μM stock solution with water, take 10 μL ...
Embodiment 3
[0152] Example 3. The RNA site-directed editing technology based on Crispr-Cas13a is used to restore the transposition function of the fission yeast retrotransposon
[0153] 1. Expression of dCas13a-hADAR2d fusion protein in fission yeast cells
[0154] Using the same method as in Example 1, the dCas13a-hADAR2d fusion protein was expressed in fission yeast cells.
[0155] 2. Expression of mutant retrotransposon TF1(G1165A) in fission yeast cells
[0156] 1. The Tf1(G1165A) gene was amplified by Over-lap PCR method
[0157] A. Amplify the 5' fragment of Tf1(G1165A) gene
[0158] With pHL414 (Professor Henry L.Levin's lab) as template, with TF1-XhoI-nmt1-P5 (ATCATCATATGGTGAGCAAGGGCGAGGAGCTGTTCACCGGGGT (SEQ ID NO: 19)) and Tf1-1165-mutant-P3 (TCAGGGTGGTCACGAGGGTGGGCTAGGGCACGGGCAGC TT0GCCGT) IDGCCG ) as primers for PCR amplification to obtain the 5' fragment of Tf1(G1165A) gene.
[0159]B. Using pHL414 as a template, Tf1-1165-mutant-P5 (ACCGGCAAGCTGCCCGTGCCCTAGCCCACCCTCGTGACCA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com