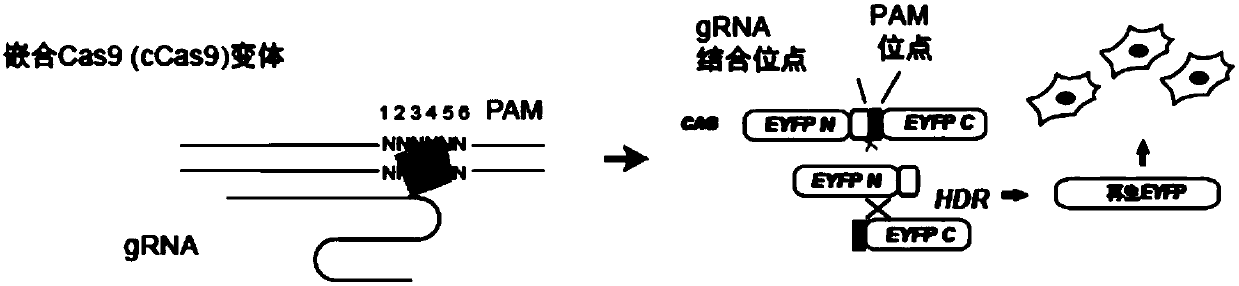
Construction of chimeric SaCas9 based on evolutionary information for enhancing and extending recognition of PAM loci
A technology for identifying regions and amino acids, applied in biochemical equipment and methods, applications, genetic engineering, etc., to achieve the effect of powerful gene editing capabilities
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0256] Example 1 Genome mining SaCas9 highly homologous protein
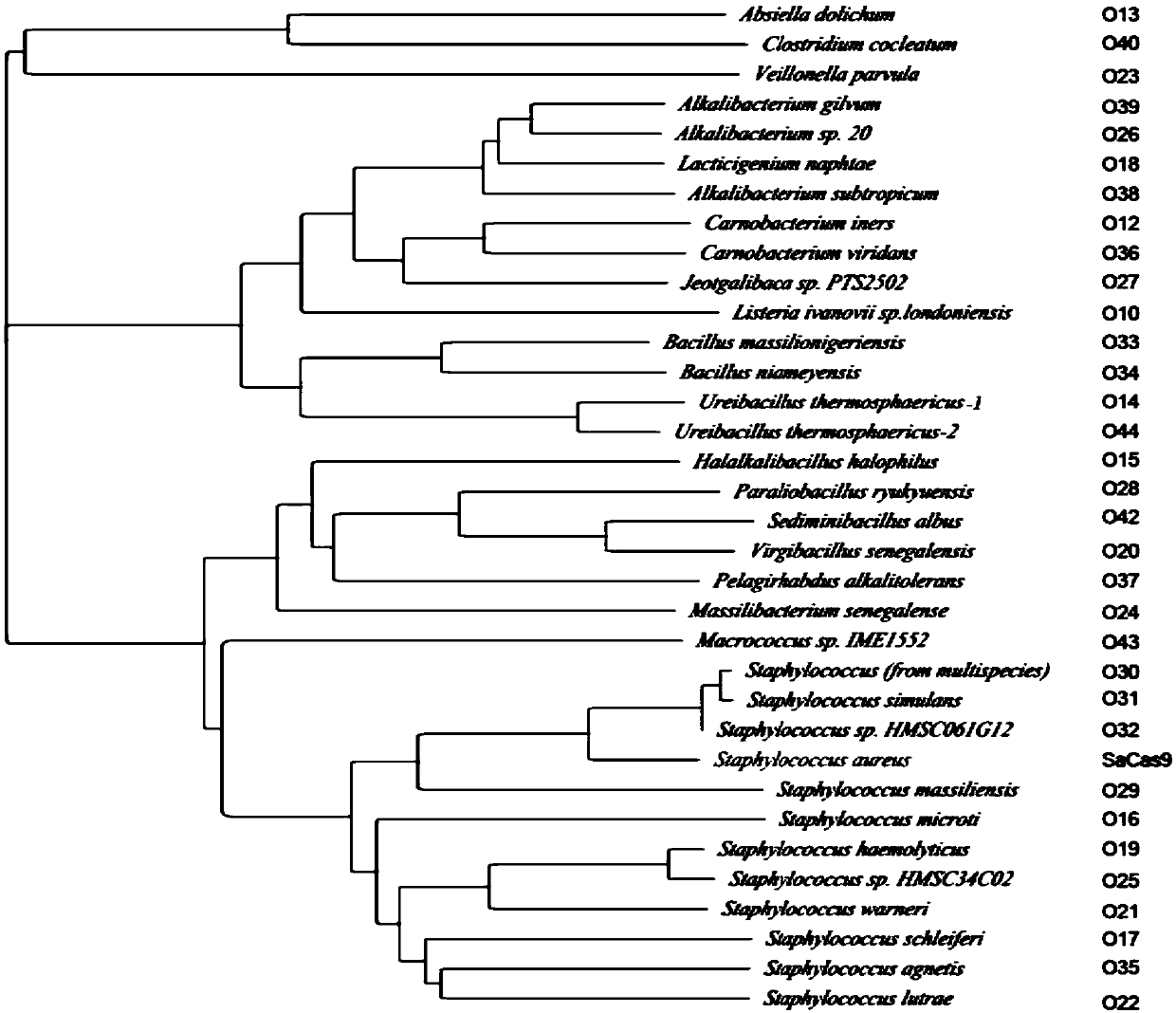
[0257] First, the inventors searched for the full-length homologous protein of SaCas9 in the NCBI database through the BLAST program. Such as figure 1 As shown (wherein, the phylogenetic tree analyzes the homologous proteins of SaCas9, wherein except SaCas9, the right column shows the numbers corresponding to Cas9), the inventors found 33 homologous proteins of SaCas9. Among them, 11 homologous proteins are from the genus Staphylococcus and have higher sequence homology with SaCas9. And found an interesting phenomenon, the Cas9 protein in bacteria from the same genus often has higher sequence homology. For different Cas9 homologous proteins, the inventors named them O+ numbers, such as O21, O22, etc.
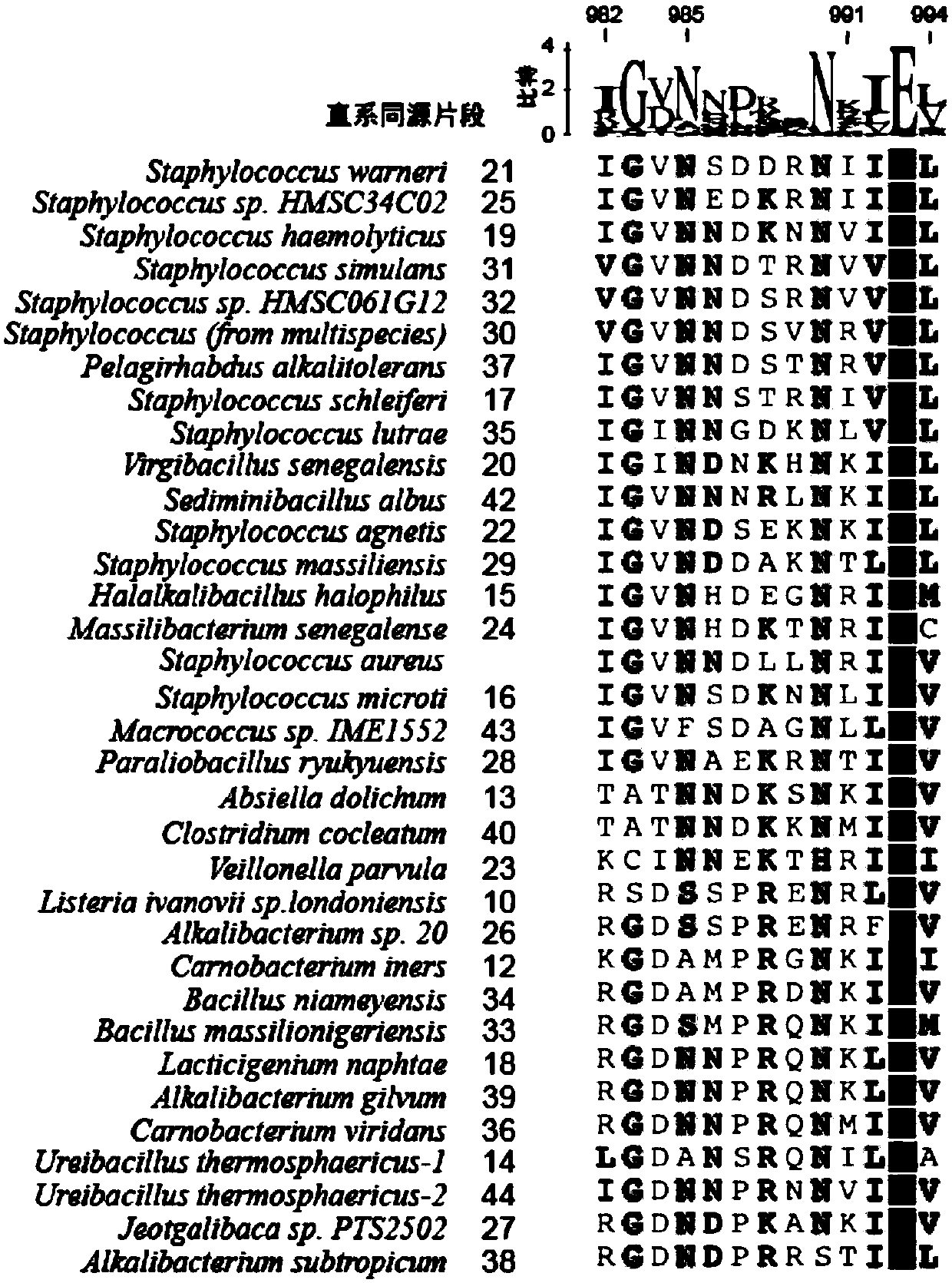
[0258] SaCas9-KKH recognizes PAM as NNNRRT, for the convenience of description, the inventor abbreviates it as RRT. Such as figure 2 As shown, through the comparison of homologous sequences, the inventors fo...
Embodiment 2
[0260] Example 2 cCas9 exhibits different PAM tendencies
[0261] The inventors and other research groups can be used to change the 4 consecutive U sequences in the gRNA backbone by changing the U in the third and fourth positions in the gRNA backbone, thereby reducing the premature termination signal due to continuous U being recognized by polymerase III . The inventors used the CIRPSRfinder program to find the CRISPR sites in bacteria corresponding to different Cas9 homologous proteins. The inventors analyzed the sequences of crRNAs corresponding to different Cas9 proteins. Interestingly, the inventors found that the crRNA direct repeat region sequences corresponding to all SaCas9 homologous proteins have sequence inconsistencies, except for the 6nt sequence at the 5' end which has sequence consistency. The inventors speculate that this is due to preventing Cas9 from targeting the cleaving DNA sequence encoding crRNA. In order to avoid sequence changes in the crRNA direct...
Embodiment 3
[0268]Next, the inventors selected mutants V17 and V42 for further analysis. In the above traversal study, the inventors found that V17 and V42 exhibited similar PAM propensity, with enhanced activity at the PAM position of RRV compared to SaCas9-KKH. The inventors first analyzed sequence comparisons at positions 982-994. Among them, V42 and V17 and SaCas9-KKH are both N at position 986, while V42 and V17SaCas9 are different at position 991. There are three amino acid differences between SaCas9-KKH and V42. Therefore, next, the inventors mutated amino acids step by step to explore the changes of PAM. Considering that position 991 is directly involved in the interaction with DNA, the inventor first mutated the position 991 of SaCas9-KKH, and mutated 991R into K. The inventor found a mutant of SaCas9-KKH (R991K), compared to SaCas9-KKH Therefore, on the basis of the 991 mutation, the inventors further mutated the amino acid D at the 987 position. Changed to N, the inventors ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com