Cancer tissue positioning method and system based on chromatin region coverage depth
A positioning method and area coverage technology, applied in the biological field, can solve the problems of low data output and poor data quality, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
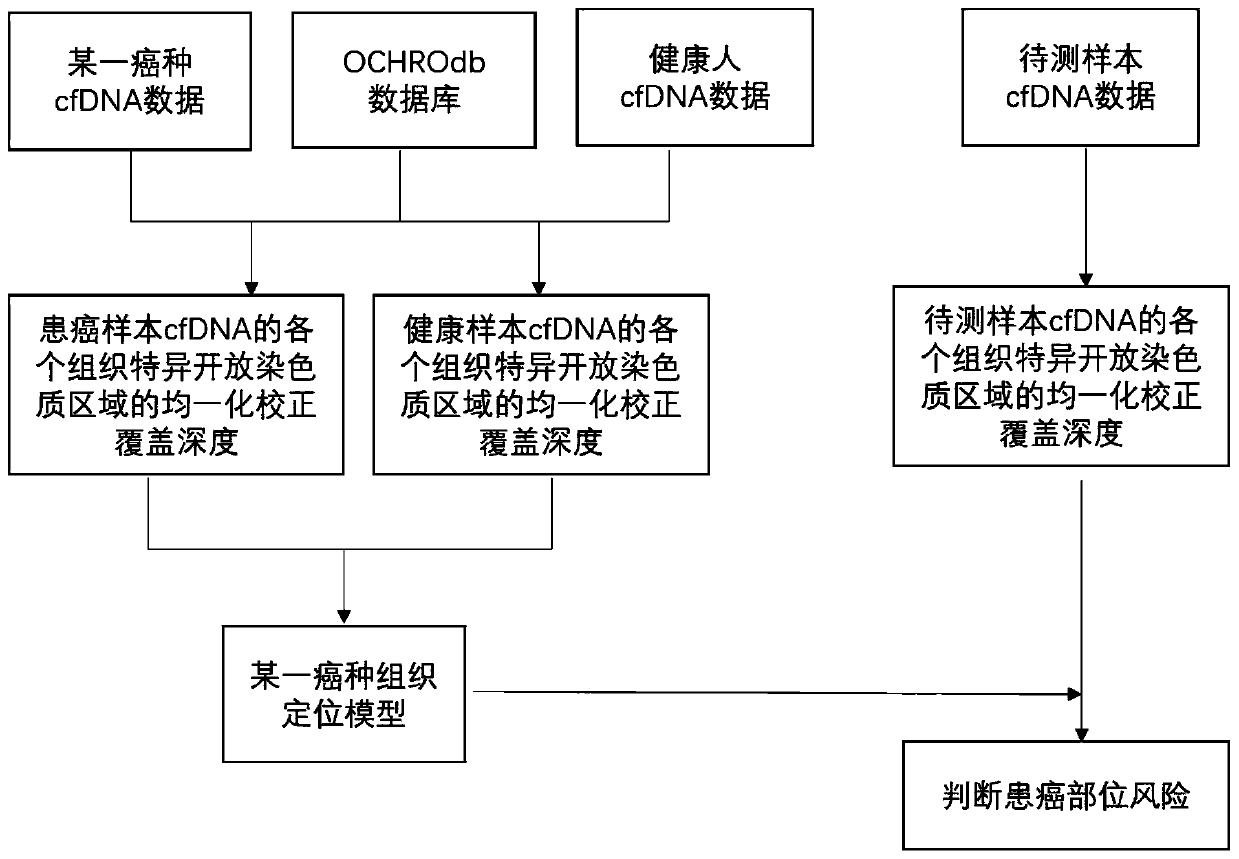
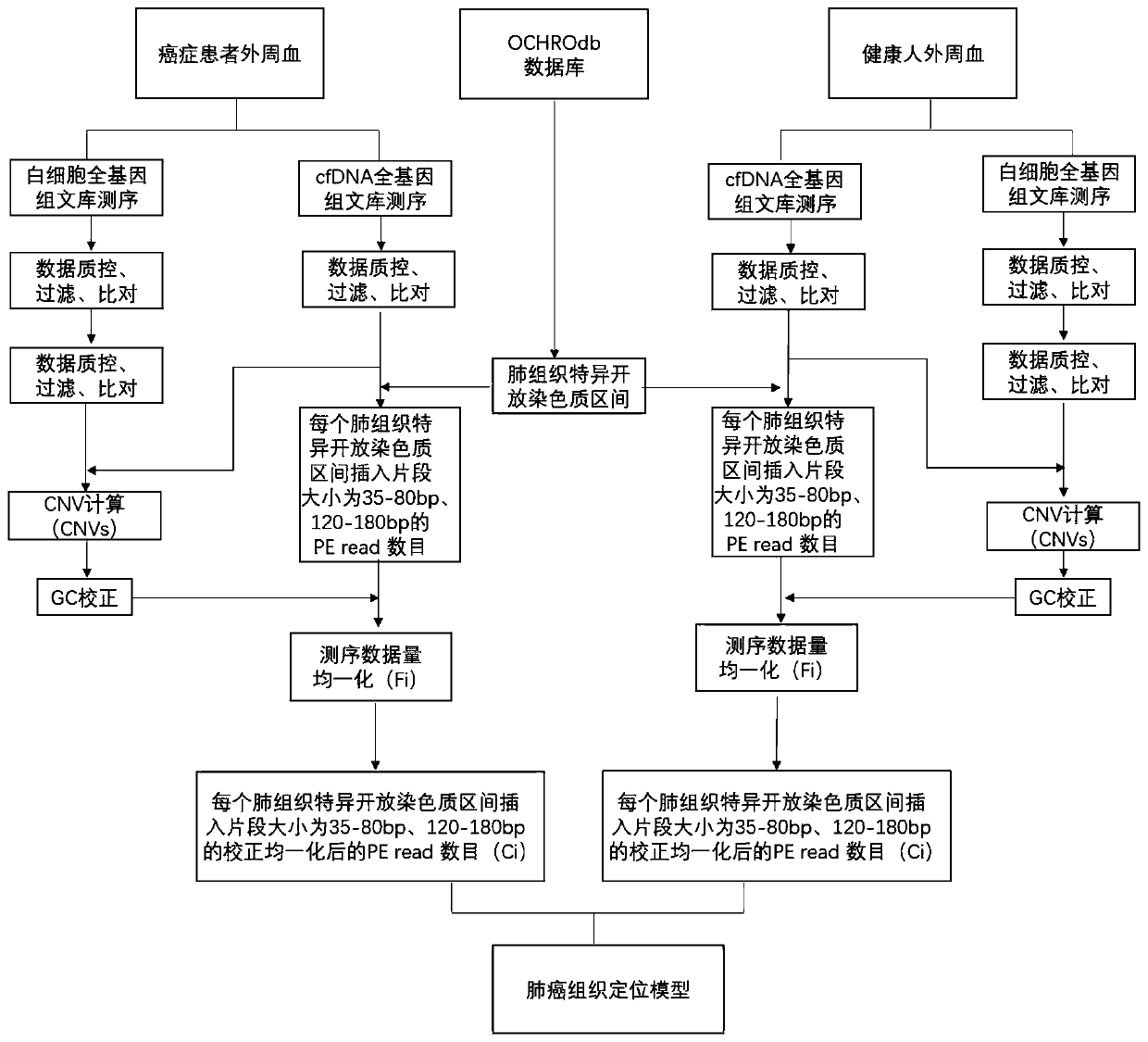
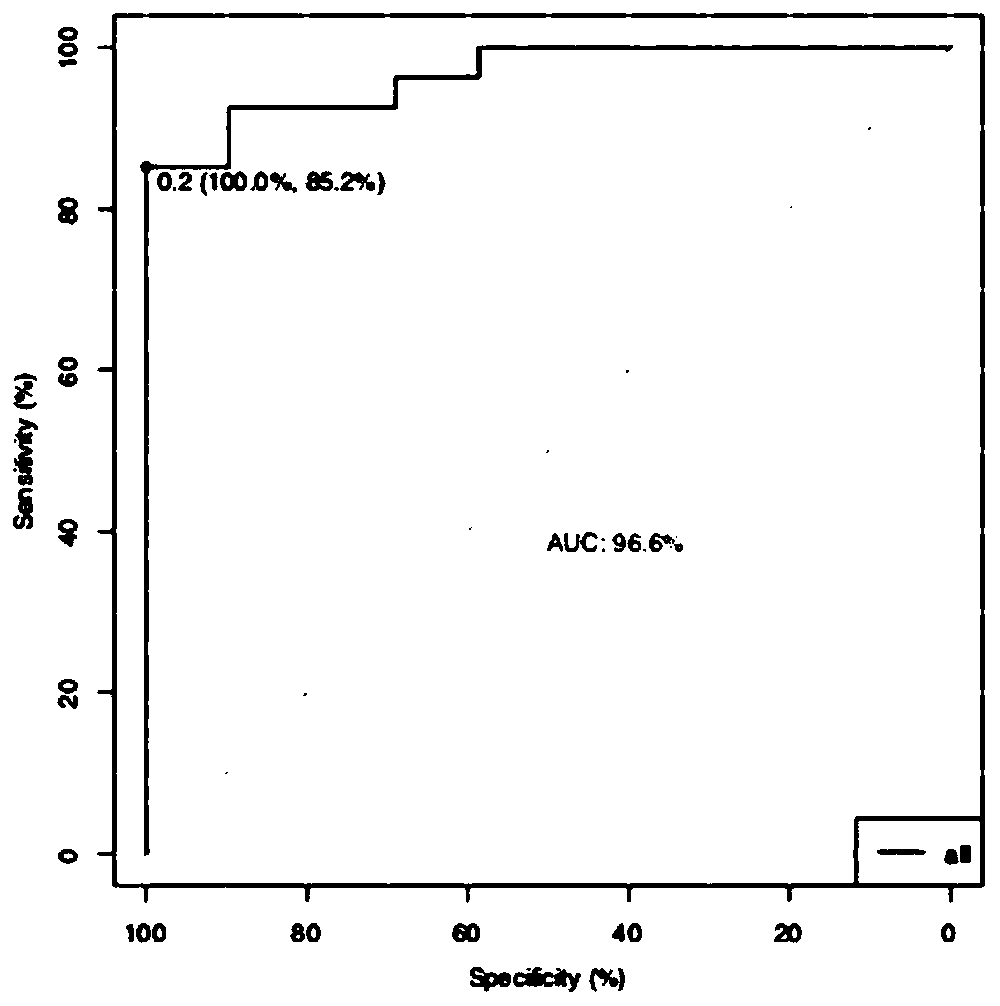
[0116] In this example, a lung cancer tissue localization model was constructed based on the coverage depth of lung tissue-specific open chromatin regions of cfDNA samples from lung cancer patients and healthy human cfDNA samples, and this model was used to predict cancer tissue localization for two samples with unknown cancer locations, as shown in figure 2 shown, including the following steps:
[0117] 1. Taking lung cancer tissue location as an example: obtain samples from 30 lung cancer patients and 30 healthy people, and build a lung cancer tissue location model, including:
[0118] 1-1 Use a 10mL Streck tube to collect the peripheral blood of the user to be tested. The blood collection volume is not less than 8mL. Immediately after blood collection, turn it upside down and mix slowly. Centrifuge the blood collection tube at 1600g for 10min at 4°C. After centrifugation, divide the upper layer of plasma into centrifuge tubes for secondary separation under the same conditi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com