Method for identifying individual intestinal flora types based on SNP
A technology of intestinal flora and intestinal bacteria, applied in the fields of genomics, instrumentation, proteomics, etc., to achieve the effect of high sensitivity, selectivity, and high detection throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
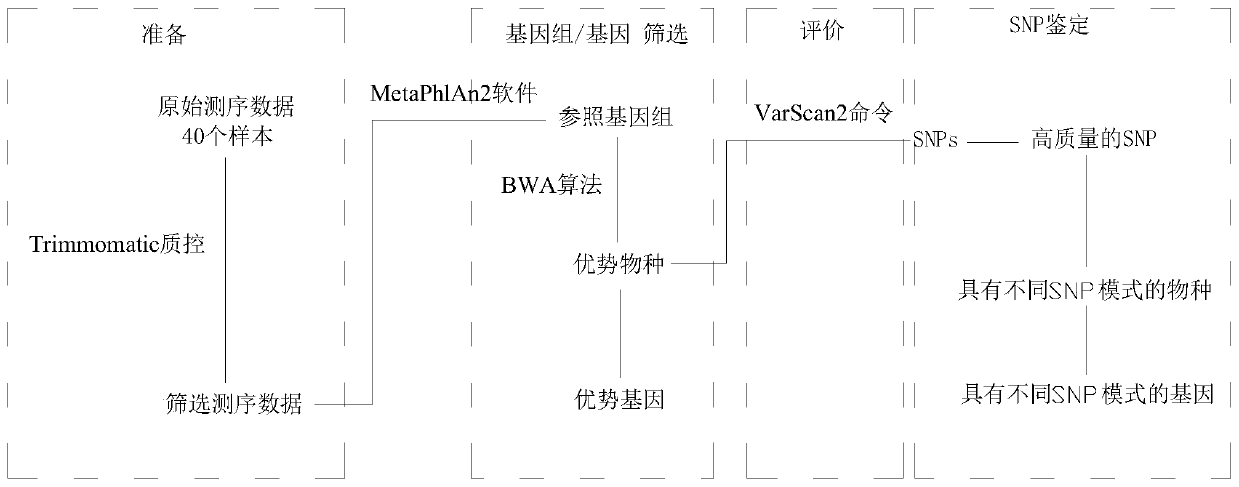
[0044] A method to identify the type of intestinal flora of an individual based on SNP, taking the identification of the type of intestinal flora of the Hadza as an example, reference Figure 1~7 , The specific identification steps are as follows:
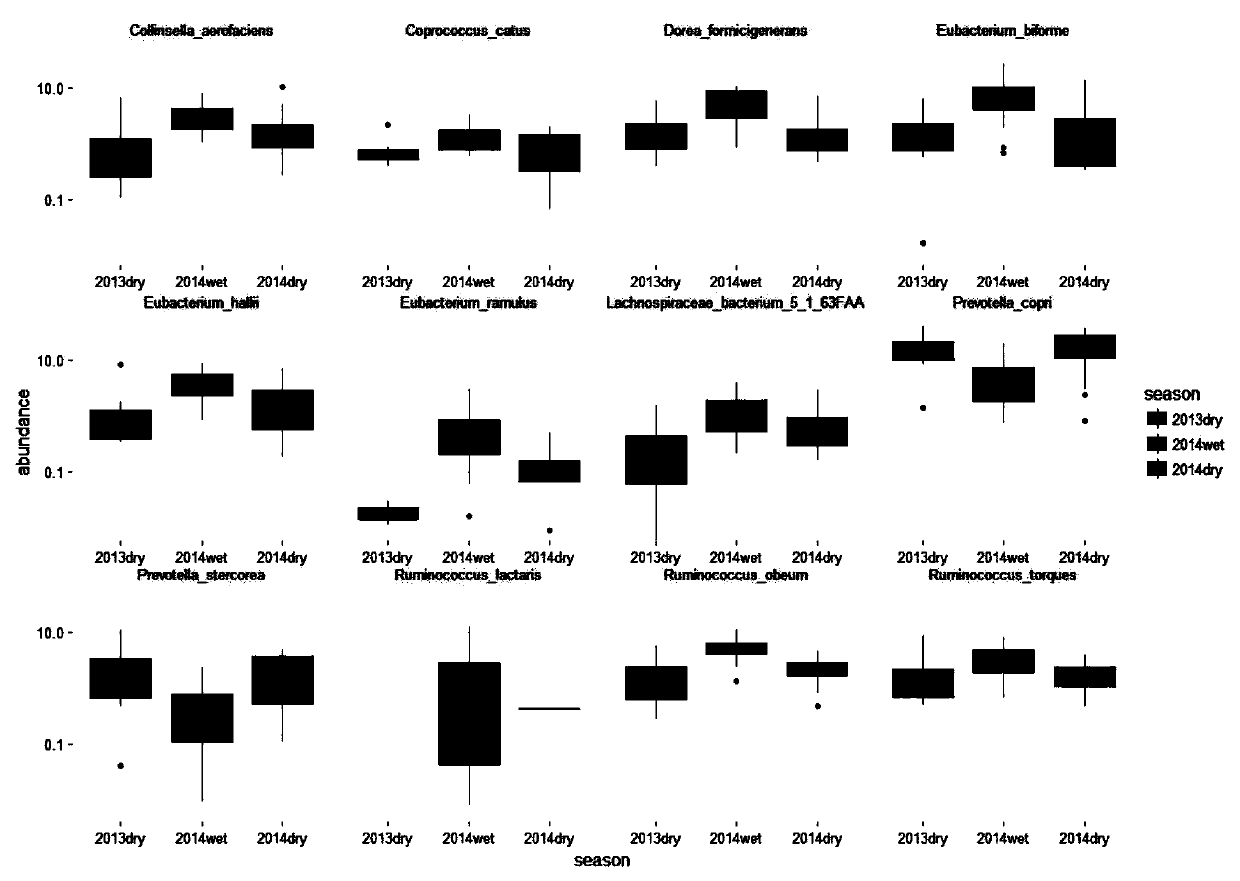
[0045] 1) Data collection: The whole genome sequencing data of Hadza human gut microbes (accession SRA582120) is obtained from the NCBI SRA database, the number of samples is 40, the data type is Illumina HiSeq 4000 single-end sequencing data, and the read length is 151bp. Among them, there were 8 samples in the dry season in 2013, 19 samples in the rainy season in 2014, and 13 samples in the dry season in 2014. Then use the fastq-dump command of sra tools to convert the obtained sra binary file into fastq, and then use the ILLUMINACLIP parameter to remove the linker. The LEADING and TRAILING parameters remove bases with a base quality of less than 5 from the head and tail of the sequence, respectively.
[0046] 2) Obtain the species c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com