Microsatellite unstable state detection kit and detection method thereof
A technology of microsatellite instability and detection kits, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., and can solve the problems of unreliable judgment results, inability to detect other polymorphic sites, Incomplete detection and other problems, to achieve accurate and reliable detection results, avoid missed judgments, and ensure sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] This embodiment provides a primer set for detecting microsatellite instability.
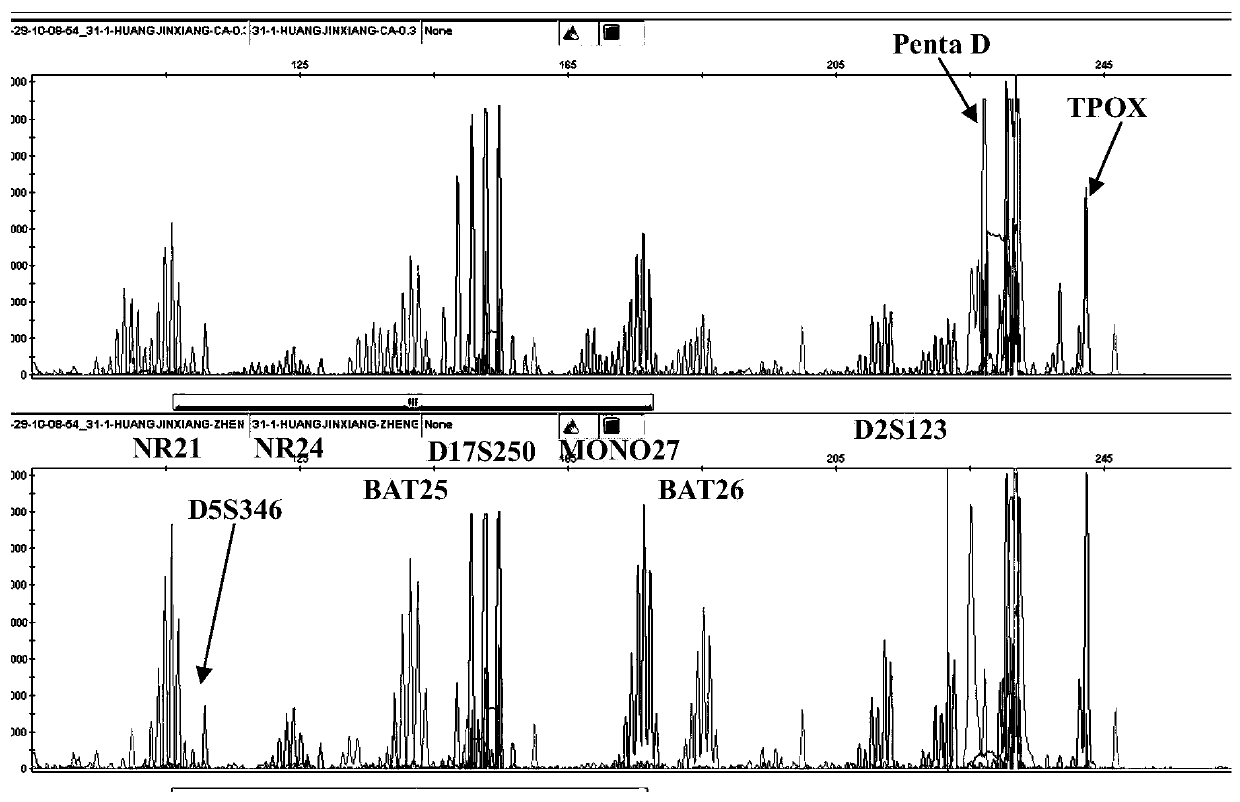
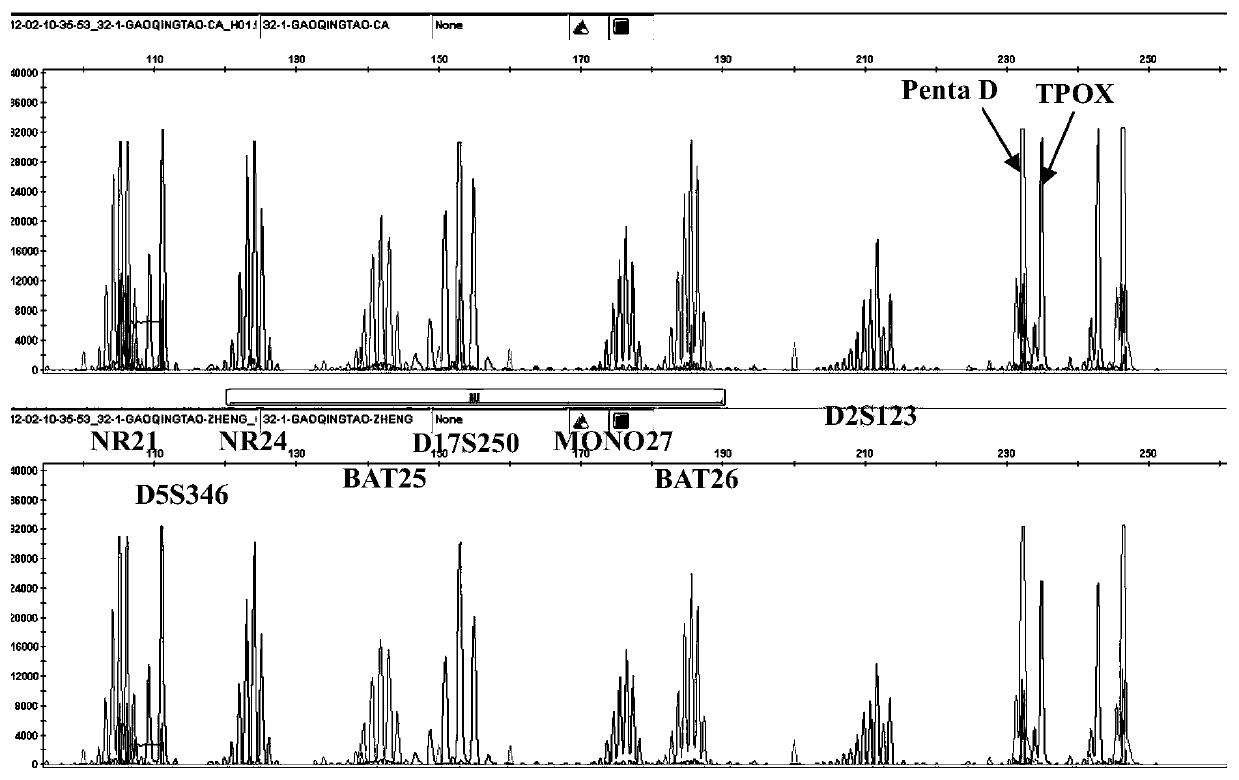
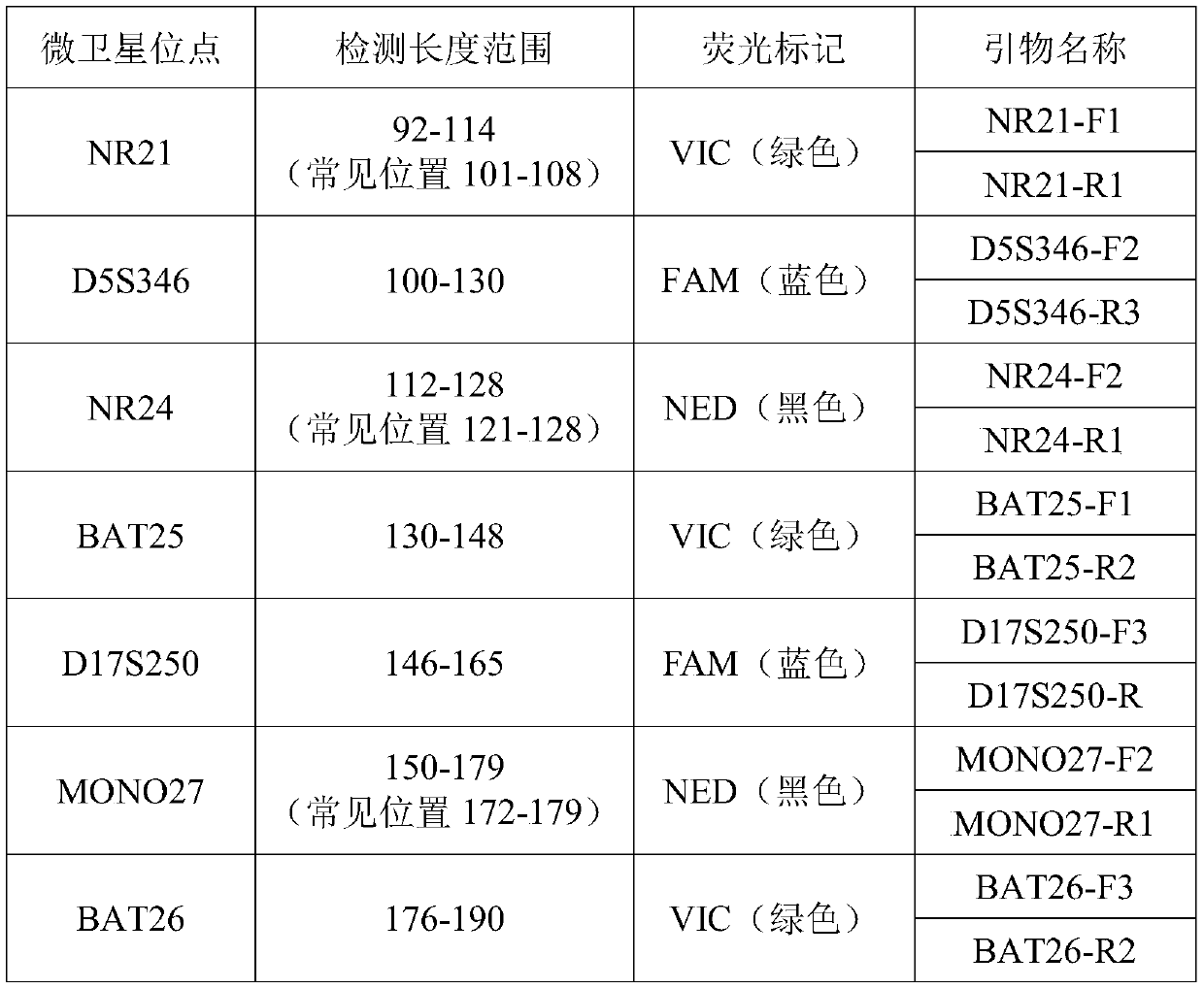
[0048] (1) This example is aimed at 5 mononucleotide repeat sites (NR21, NR24, BAT25, MONO27 and BAT26), 3 dinucleotide repeat sites (D5S346, D17S250 and D2S123) and control pentanucleotide repeats Primers were designed for the site (Penta D) and the control tetranucleotide repeat site (TPOX). The details of the 10 detection sites are as follows:
[0049]
[0050]
[0051] (2) Design of multiple fluorescently labeled primers
[0052] NR21-F1: 5'-VIC-TAAATGTATGTCCCCTGG-3' (SEQ ID NO.1)
[0053] NR21-R1: 5'-TTACATACAGAGGGGACCTG-3' (SEQ ID NO.2)
[0054] D5S346-F2: 5'-FAM-AATGAGTGAGATCACTATTTAGCCC-3' (SEQ ID NO.3)
[0055] D5S346-R3: 5'-TCGTCTATTCTGTCATAATGATCAA-3' (SEQ ID NO.4)
[0056] NR24-F2: 5'-NED-GACTTAAAATGGAGGACTGAGGTTT-3' (SEQ ID NO.5)
[0057] NR24-R1: 5'-AACACGGTAACGTAAGGTTGGACCC-3' (SEQ ID NO.6)
[0058] BAT25-F1: 5'-VIC-CGCCTCCAAGAATGTAATGT-3' (SEQ ID NO.7)
[0059]...
Embodiment 2
[0077] For the detection method of microsatellite instability provided in this embodiment, the sample to be tested can be any form of sample that can extract nucleic acid, including but not limited to whole blood, serum, plasma and tissue samples; tissue samples include but not limited to paraffin-coated Buried tissue, fresh tissue, and frozen sections.
[0078] S1. Sample genomic DNA extraction
[0079] Two patients with colorectal cancer were taken, one of which was detected as MSI-H by MMR immunohistochemistry, and the other was detected as MSS by MMR immunohistochemistry, and the cancerous tissue samples and normal tissue samples (peripheral blood or cancerous tissue 3cm around), make paraffin-embedded tissue sections with a thickness of 5-20μm (make sure the section contains 9mm 2 Tissue blocks of the above size), use the paraffin-embedded tissue DNA extraction kit from TIANGEN Company to extract genomic DNA from the paraffin-embedded tissue sections of the two samples, ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com