Multi-animal-derived adulteration identification method based on whole genome sequencing technology
A whole-genome sequencing, animal-derived technology, applied in the field of biological analysis, can solve the problems of amplification bias, increase the complexity of data integration, and the difficulty of direct quantification of template DNA molecules, and achieve the effect of wide application prospects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] 1. Mock sample preparation, library construction and next-generation DNA sequencing
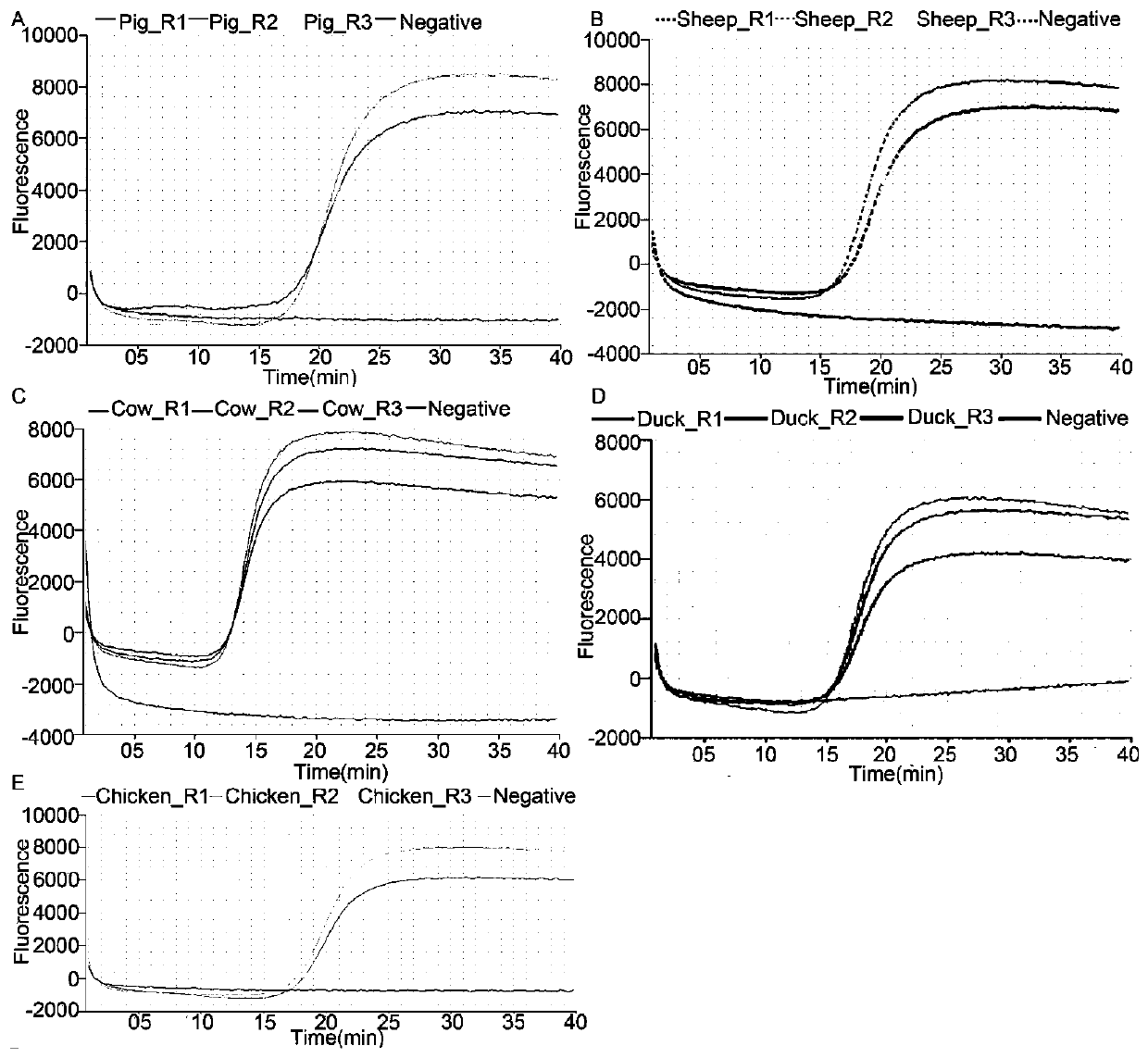
[0038] The simulated samples were prepared from the meat of 15 kinds of animals, including duck, cow, camel, dog, horse, chicken, mouse, ferret, nutria, raccoon dog, rabbit, sheep, rat, pig and fox, among which Duck, beef, chicken and pork were purchased from local markets in Beijing, China, and other meats were collected and identified by the Xiamen Customs Technology Center from January 2017 to December 2018. The meat samples of the above species were selected mainly considering that some of them have great economic significance, and some of them often appear in adulterated foods.
[0039] Fresh meat samples were immediately frozen and stored in a -80°C freezer until use. The samples were pooled in two ways, a pooled sample containing an equal mass pool from 15 meats, hereinafter referred to as "M15"; M15 had three replicates, labeled "R1", "R2" and "R3". Another mixed sample conta...
Embodiment 2
[0117] The quantitative analysis of two kinds of mixtures of embodiment 2
[0118] The analysis in Example 1 shows that WGS plus mitochondrial genomes can qualitatively identify taxon components in mixed samples. To determine how quantitative the method is, a series of mock samples were prepared using different proportions of porcine and chicken material:
[0119] The M2 mixed sample in Example 1 was used for processing, and the DNA extraction, library construction, DNA sequencing and DNA analysis methods were the same as those of the M15 sample, and the DNA sequencing results are shown in Table 1. Such as Figure 5 As shown, the proportion of the unique sequence compared to the pig reassembled mitochondrial genome in the mixed sample NGS data (raw sequencing sequence) (3 groups mean) and the weight ratio of the mixed sample ( Figure 5 The correlation coefficient between A) is R 2 = 0.978. Similarly, the correlation coefficient between the proportion of the unique sequenc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com