Method for detecting tumor gene mutation through combination of sequence specific blocking agent (SSHB) and specific PCR program
A program detection and tumor gene technology, which is applied in the measurement/testing of microorganisms, biochemical equipment and methods, etc., can solve the problems of low amplification efficiency, large △CT value, weakened mutation detection rate, etc., and improve the detection rate. The effect of increasing the output rate and increasing the binding rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
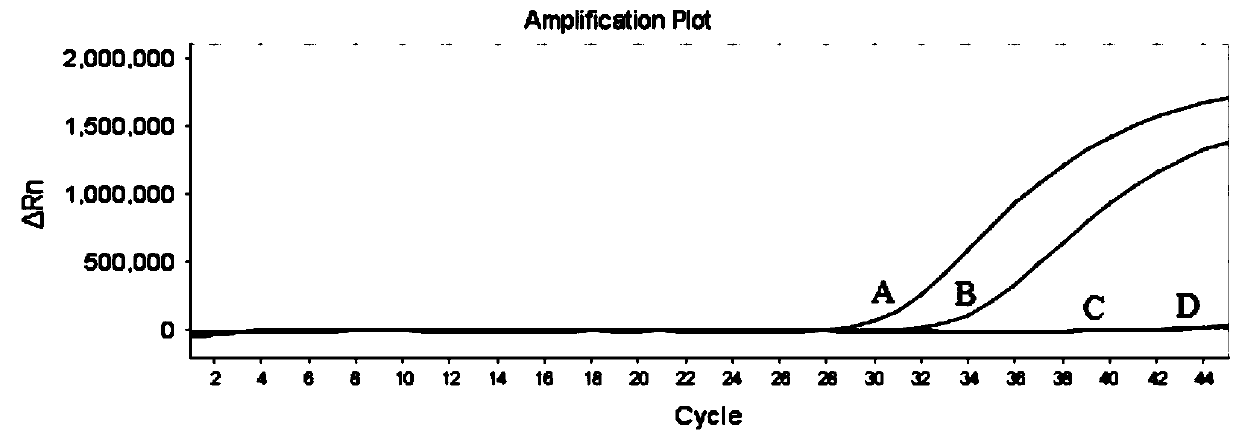
[0037] Example One-thousandth mutation detection of gene locus BRAF (V600E) for tumor-targeted drugs
[0038] 1. Design of primers, probes and SSHB
[0039] Open the UCSC website, enter the BRAF gene, download the entire gene sequence of BRAF, and then locate the specific gene locus according to the RS113488022 of BRAF (V600E), and select the bases of 100 bp upstream and downstream of the BRAF (V600E) locus to analyze the SNP; open NCBI's SNP database, input RS113488022, you can query the SNP population frequency of this site, and at the same time, you can query all the SNP sites upstream and downstream of the site in the Variation Viewer, and query the SNP population frequency of these sites one by one.
[0040] (1) Primer and probe design
[0041] Primer design: SNP analysis is performed on the upstream and downstream sequences of the mutation site to ensure that the ARMS primers do not contain high-frequency SNP sites, and other upstream and downstream primers are also sel...
Embodiment 2
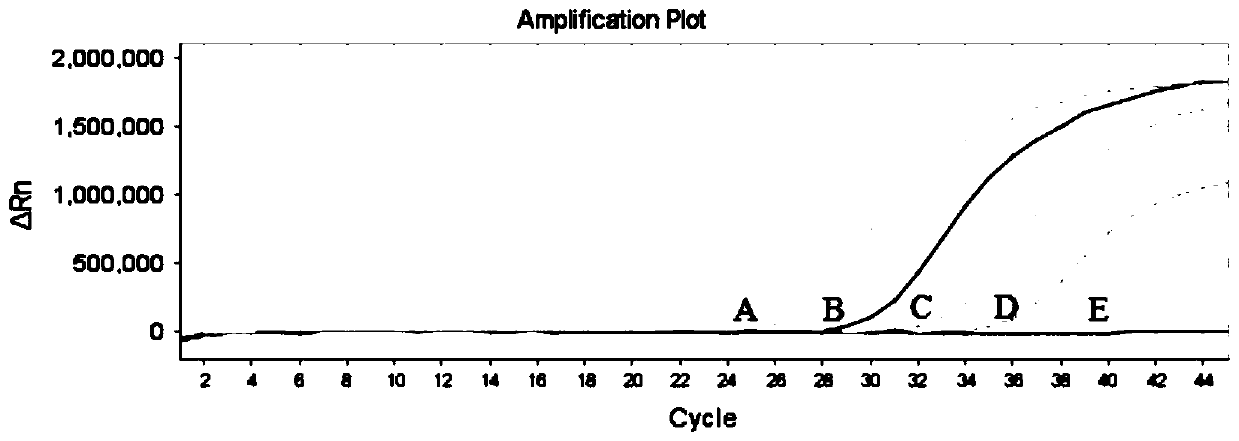
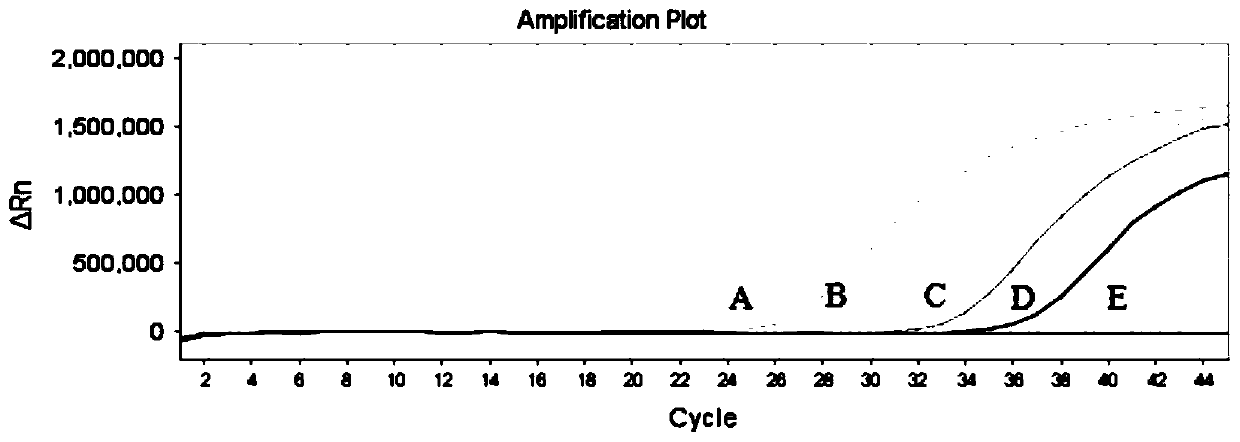
[0066] Example 2 Detection of one-thousandth mutation of tumor-targeted drug gene locus KRAS (G12A)
[0067] 1. Design of primers, probes and SSHB
[0068] Open the UCSC website, enter the KRAS gene, download the entire gene sequence of KRAS, and then locate the specific gene locus according to the RS121913529 of KRAS (G12A), and select the bases of 100 bp upstream and downstream of the KRAS (G12A) site to analyze the SNP; open NCBI's SNP database, input RS121913529, you can query the SNP population frequency of the site, and at the same time, you can query all the SNP sites upstream and downstream of the site in the Variation Viewer, and query the SNP population frequency of these sites one by one.
[0069] (1) Primer and probe design:
[0070] Primer design: SNP analysis is performed on the upstream and downstream sequences of the mutation site to ensure that the ARMS primers do not contain high-frequency SNP sites, and other upstream and downstream primers are also selecte...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com