Filling method and device for genotype data missing and server
A technology for missing data and missing genes, applied in genomics, instrumentation, proteomics, etc., can solve the problems of low filling efficiency and high error rate of gene filling values
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
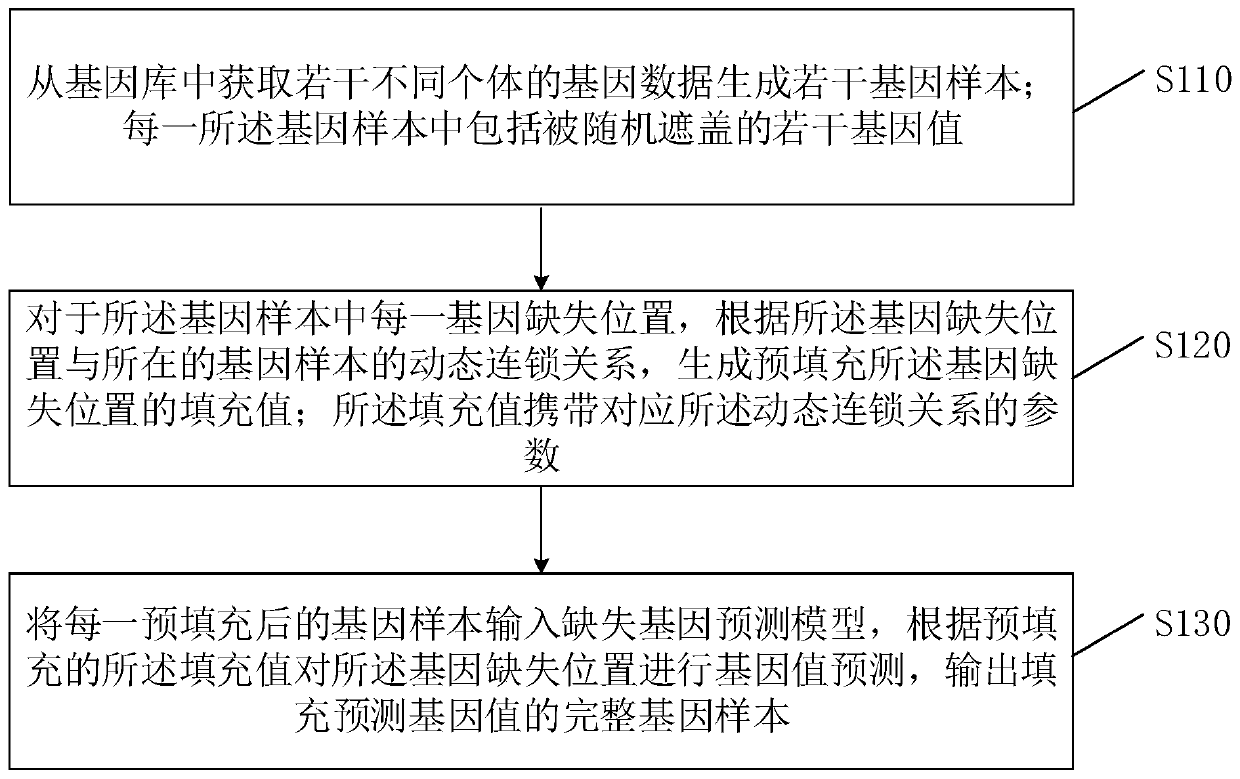
[0047] Such as figure 1 Shown is a schematic flow chart of the method for filling missing genotype data provided in the first embodiment of the present invention. This embodiment can be applied to the application scenario of predicting and filling missing gene values in genetic data. The method can be executed by a processor in a filling device with missing genotype data. The device can be a server, a smart terminal, a tablet, or a PC. Etc.; In the embodiments of this application, the filling device with missing genotype data is used as the execution subject for description. The method specifically includes the following steps:
[0048] S110. Obtain genetic data of several different individuals from the gene bank to generate several gene samples; each of the gene samples includes several gene values that are randomly covered;
[0049] In the actual sequencing of genetic samples, some genetic data in the samples will be missing in the process of processing the genetic samples t...
Embodiment 2
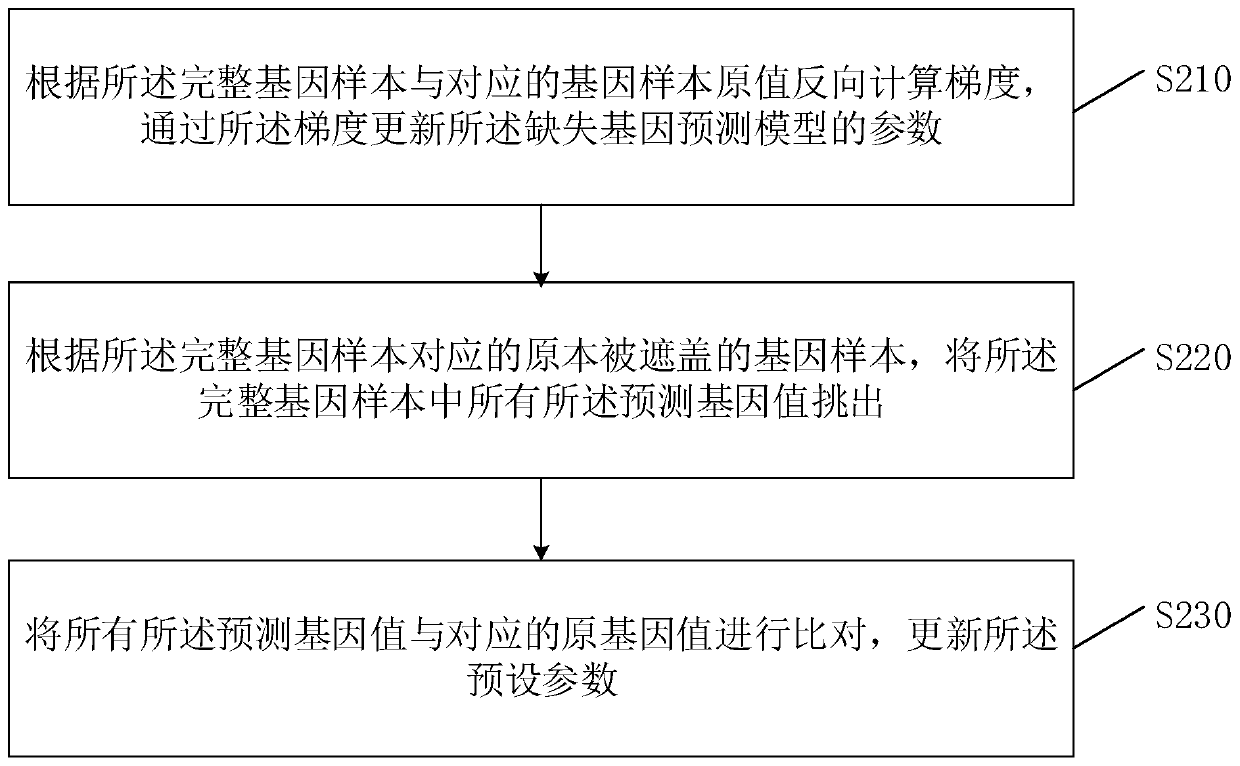
[0074] Such as image 3 Shown is a schematic flow chart of the method for filling in missing genotype data provided in the second embodiment of the present invention. On the basis of the first embodiment, this embodiment also provides a process of optimizing the parameters carried by the values and the parameters of the missing gene prediction model in the method for filling missing genotype data, thereby further improving the accuracy of predicting gene values. The method specifically includes:
[0075] S210: Calculate a gradient in reverse according to the original value of the complete gene sample and the corresponding gene sample, and update the parameters of the missing gene prediction model through the gradient;
[0076] Due to the filling method of missing genotype data, the process of predicting and filling the test gene samples with missing gene data includes two parts. The first part is that for each missing position of the gene to be tested, based on the dynamic linka...
Embodiment 3
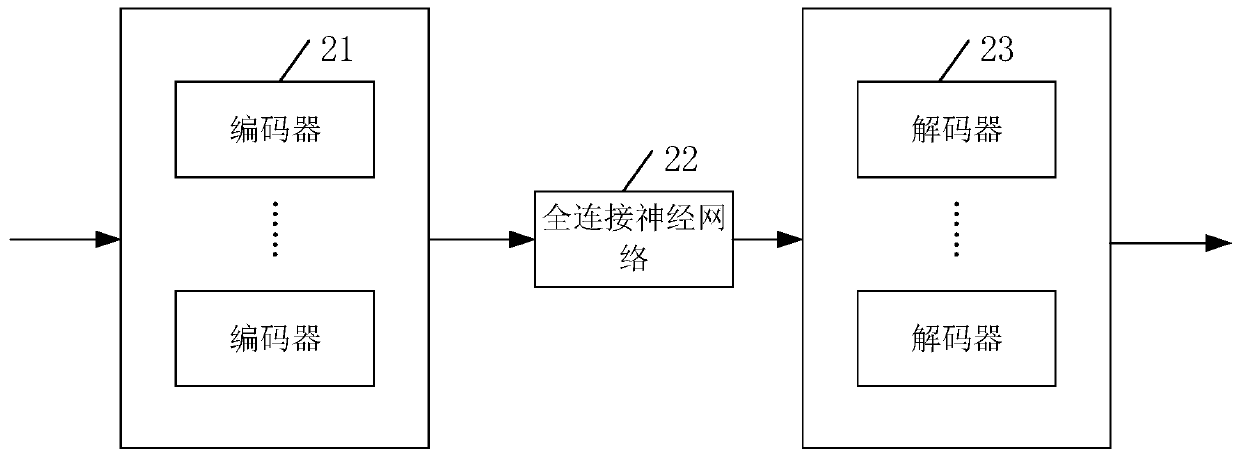
[0085] Such as Figure 4 Shown is the filling device with missing genotype data provided in the third embodiment of the present invention. On the basis of Embodiment 1 or 2, an embodiment of the present invention also provides a detection device 4, which includes:
[0086] The gene sample generating module 401 is configured to obtain gene data of several different individuals from the gene bank to generate several gene samples; each of the gene samples includes several gene values that are randomly covered;
[0087] In an implementation example, the genetic data of several different individuals is obtained from the gene bank to generate several gene samples; when each of the gene samples includes several gene values that are randomly covered, the gene sample generation module 401 includes:
[0088] The intercepting unit is used to obtain genetic data of several different individuals from the gene bank, and intercept the genetic data into several genetic samples with the same leng...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com