Detection method for helicobacter pylori 23S rRNA (Ribosomal Ribonucleic Acid) gene drug-resistant mutation, and application of detection method
A technology of Helicobacter pylori and a detection method, which is applied in the field of detection of drug resistance mutations of Helicobacter pylori 23SrRNA gene, can solve the problems of inability to detect polyclonal conditions, short read length of second-generation sequencing sequences, and inability to distinguish drug resistance mutations, etc. Drug preparation efficiency and the effect of improving resolution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
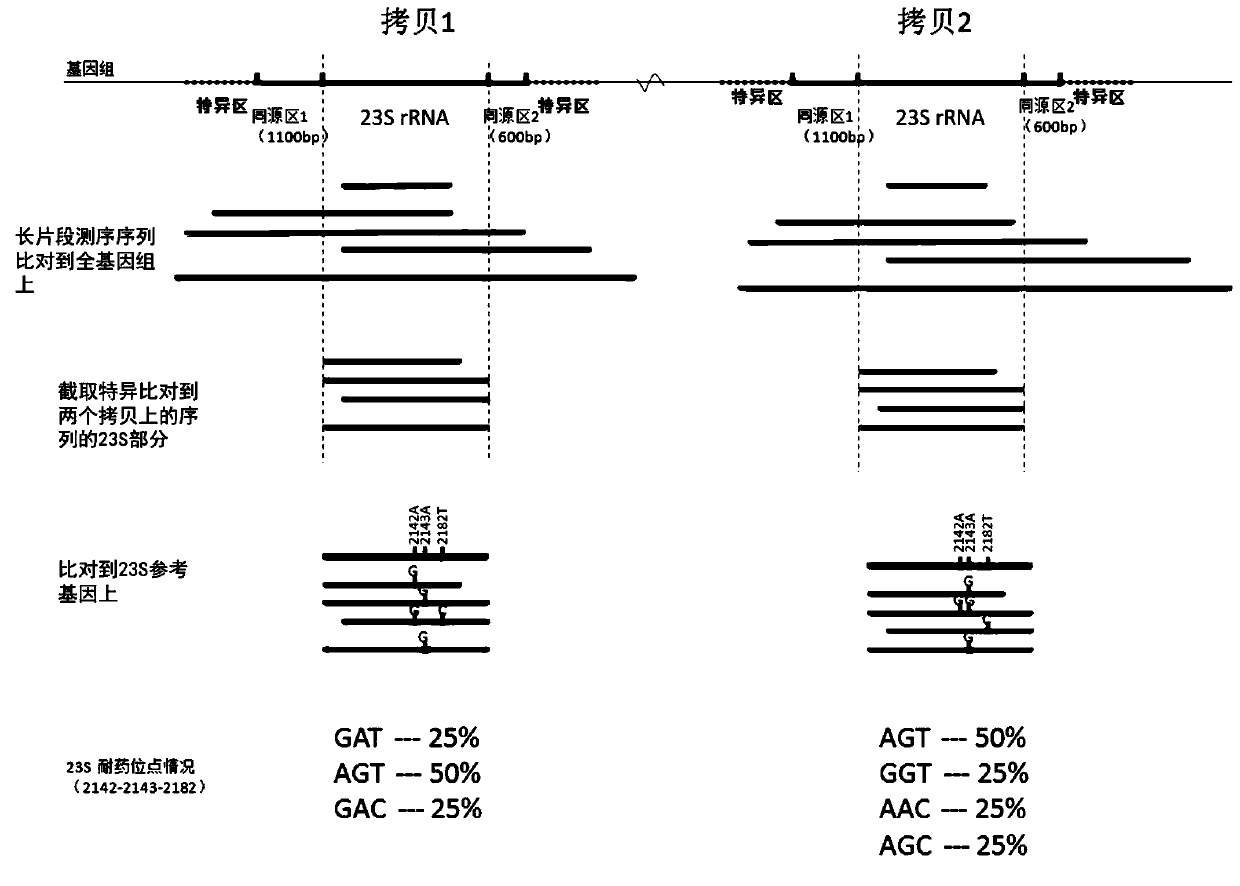
[0063] (1) Genomic DNA was extracted from the Helicobacter pylori samples to be detected from gastric biopsies of patients with gastritis in the First Affiliated Hospital of Wenzhou Medical University, and then the library was constructed, and then sequenced using the PromethION sequencer from Oxford Nanopore. After the sequencing was completed The software recommended by the sequencer company was used to split the data to obtain the long-fragment sequencing data of the sample. In order to obtain a more accurate genome and to verify the results of the experimental protocol, the MGI sequencer manufactured by BGI was also used for next-generation sequencing.
[0064] (2) Use the long-segment sequencing data and the second-generation data to assemble the complete genome of the sample. Along the direction of the genome, first search for the specific sequence GGCAAGACGG and GACCC to find the copy of the 23S rRNA gene in the same direction as the genome, and then search in the revers...
Embodiment 2
[0069] (1) Genomic DNA was extracted from the Helicobacter pylori sample (13478) to be detected from the gastric biopsy of another patient with gastritis in the First Affiliated Hospital of Wenzhou Medical University, and then the library was constructed, and then the PromethION sequencer of Oxford Nanopore was used for the sequencing Sequencing, after the sequencing is completed, use the software recommended by the sequencer company to split the data to obtain the long-fragment sequencing data of the sample. In order to obtain a more accurate genome and to verify the results of the experimental protocol, the MGI sequencer manufactured by BGI was also used for next-generation sequencing.
[0070] (2) Use the long-segment sequencing data and the second-generation data to assemble the complete genome of the sample. Along the direction of the genome, first search for the specific sequence GGCAAGACGG and GACCC to find the copy of the 23S rRNA gene in the same direction as the genom...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com