CRISPR-Cas9-specific method for knocking out pig Fah and Rag2 genes
A specific, double-gene technology, applied in the field of gene editing, to achieve the effect of reasonable design and intuitive screening
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Example 1 Selection and design of Sus scrofa (pig) Fah and Rag2 gene sgRNA target sequences
[0051] (1) Method
[0052] 1. Selection of sgRNA target sequences for Fah and Rag2 genes
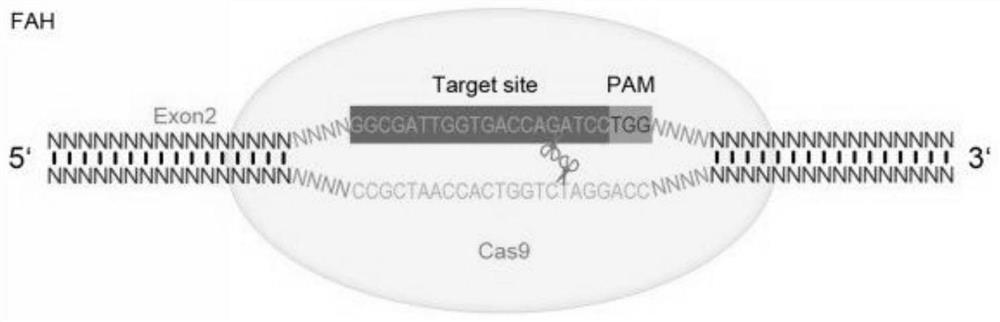
[0053] To find sgRNA target sequences in the exon coding regions of Fah and Rag2 genes, the principles are as follows:
[0054] (1) The general sequence formula is 5'-N(20)NGG-3', where N(20) represents 20 consecutive bases, and each N represents A or T or C or G, and targets that meet the above rules The sequence is on the sense or antisense strand;
[0055] (2) For the Fah gene, select the 5 exon coding region sequences near the N-terminal of the gene Fah, the target sequence can be located at the 5 exon coding regions at the N-terminal of the Fah gene, or a part of the target sequence is located at the N-terminal of the Fah gene 5 exons, the rest spans the junction with the adjacent intron and is located in the adjacent intron; the cutting of such coding region sequence will cause t...
Embodiment 2
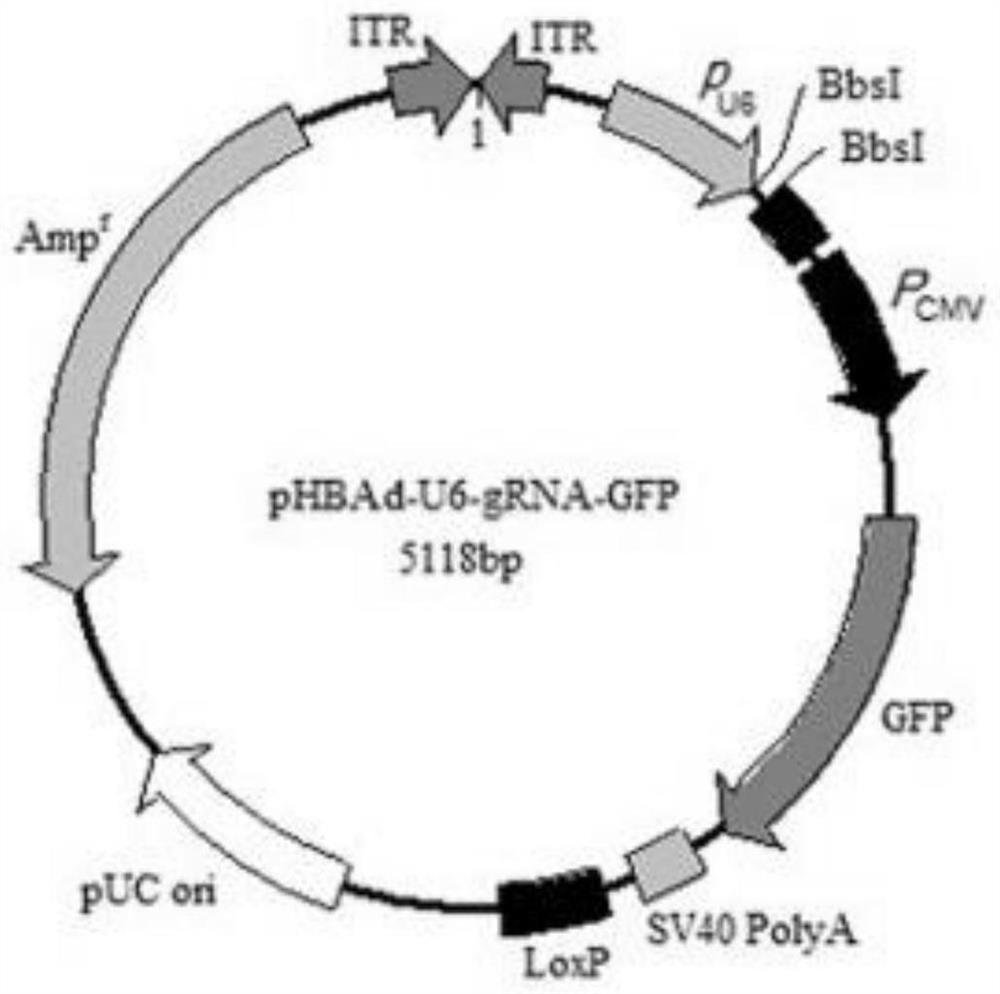
[0075] Example 2 Construction of sgRNA expression vectors of Fah gene and Rag2 gene and adenovirus packaging
[0076] 1. Synthesis of DNA Inserts
[0077] (1) Synthesize the Fah-sgRNA and Rag2-sgRNA forward and reverse oligonucleotide sequences designed above
[0078] The oligonucleotide sequence can be specifically synthesized by a commercial company (such as Invitrogen) according to the provided sequence. The forward oligonucleotide sequence and reverse oligonucleotide sequence corresponding to No. 17 target sequence in Table 1 are as follows:
[0079] 5'-CACCGGCGATTGGTGACCAGATCC-3' (SEQ ID NO.91):
[0080] 5'-AAACGGATCTGGTCACCAATCGCC-3' (SEQ ID NO. 92).
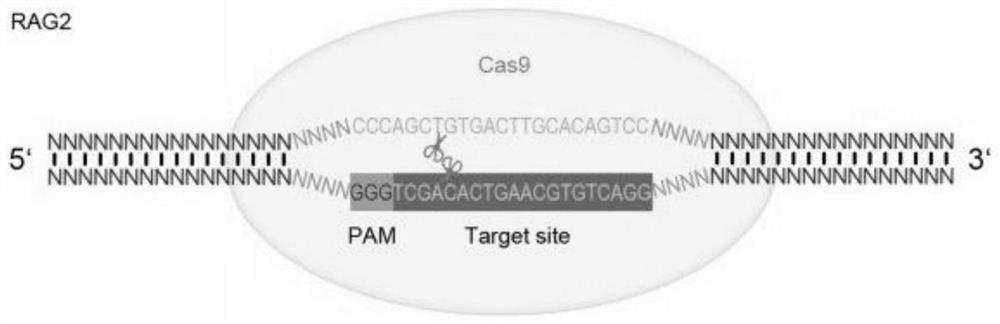
[0081] The forward oligonucleotide sequence and reverse oligonucleotide sequence corresponding to No. 1 target sequence in Table 2 are as follows:
[0082] 5'-CACCGGACTGTGCAAGTCACAGCT-3' (SEQ ID NO.93);
[0083] 5'-AAACAGCTGTGACTTGCACAGTCC-3' (SEQ ID NO. 94).
[0084] The gene knockout pattern after the sgRNA corresp...
Embodiment 3
[0136] Example 3 Obtaining a pseudotyped lentivirus expressing Cas9
[0137] 1. Material preparation
[0138] Virus packaging system: three-plasmid system, pSPAX2, pMD2G, pHBLV-Cas9-Puro (purchased from Hanbio, Figure 5 ); culture packaging cell line HEK293T cells (purchased from ATCC); DMEM medium, Opti-MEM medium and fetal bovine serum FBS (purchased from Gibco); Lipofectamine2000 (purchased from Invitrogen); HEK293T cells were cultured in 5% CO2 containing In a 37°C culture environment, the culture medium is DMEM medium containing 10% FBS.
[0139] 2. Transfection and Viral Packaging
[0140] Day 1: Passage the packaging cell line HEK293T to T75, about 30% confluence;
[0141] The next day: When HEK293T reaches 50%-60% confluence, transfect according to the following recipe:
[0142] Prepare Mixture 1, containing:
[0143] pSPAX2 10 μg
[0144] pMD2G 10 μg
[0145] pHBLV-Cas9-Puro vector 10 μg
[0146] Opti-MEM: 500 μL
[0147] Prepare Mixture 2, containing:
[0...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com