HRM primer pair, kit and method for rapidly identifying feline enteric coronavirus and feline infectious peritonitis viruses
A technology of peritonitis virus and coronavirus, applied in the field of molecular biology detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Embodiment 1. is used to distinguish the design of the HRM primer of feline enteric coronavirus and feline infectious peritonitis virus
[0037] 1.1 Virus isolation and cultivation and nucleic acid extraction
[0038] FIPV was isolated and cultured in our laboratory, and FECV nucleic acid was extracted from clinical samples. Nucleic acid RNA was extracted from cat ascites and intestinal feces samples according to the instructions of the RNA extraction kit from TIANGEN.
[0039] 1.2 Design of HRM primers
[0040] The full-length sequences of FECV and FIPV were downloaded from NCBI (Genebank accession numbers are KY566209 and KC461237.1, respectively) for comparison. Attempts to design primers on the commonly used conserved gene S or N gene were unsuccessful. Finally, a pair of primers were designed on a relatively conserved sequence on the 7b gene (between 29 000 bp and 29 500 bp) (primer sequences are shown in Table 1). The pair of primers can specifically amplify a...
Embodiment 2
[0045] Example 2. Establishment of the HRM Response for Distinguishing Feline Enteric Coronavirus and Feline Infectious Peritonitis Virus
[0046] Using the extracted nucleic acid as a template, use the upstream and downstream primers in Table 1 to amplify. A total of 25 μl of RT-PCR reaction system: RNA 1 μl, PrimerScript 1step Enzyme 1 μl, 2×step Buffer 12.5 μl, RNase Free ddH 2 O 5.5 μl, upstream primer 1 μl, downstream primer 1 μl, fluorescent dye LC GREEN 1 μl. The working concentration of upstream and downstream primers was 10 μmol / L.
[0047] The RT-PCR reaction program was 50°C for 30 minutes and 95°C for 2 minutes. Pre-denaturation at 95°C for 30s; 35 cycles at 95°C for 10s, 55°C for 20s, 72°C for 30s, 72°C for 5min, 4°C for 30min. In the HRM analysis, it is 75°C-88°C, with a heating rate of 0.1°C / step, collecting fluorescence every 0.05s for melting curve analysis.
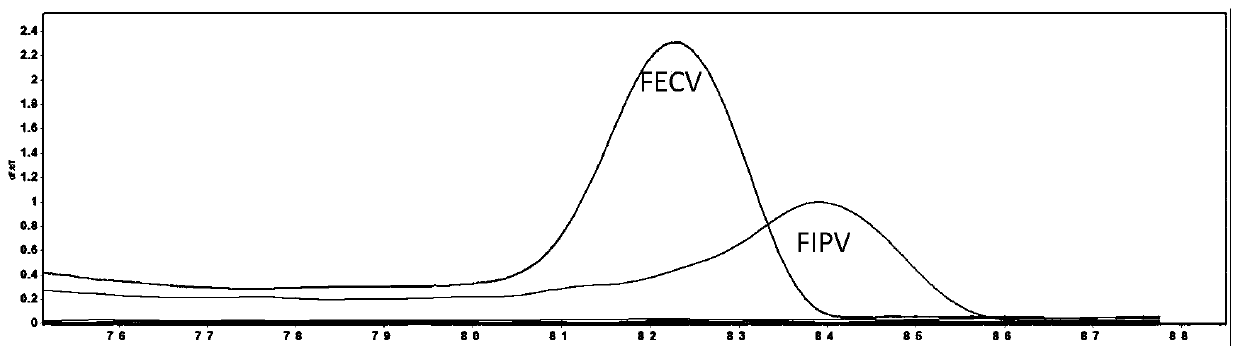
[0048] The results show that the present invention has successfully established a HRM method for si...
Embodiment 3
[0049] Embodiment 3, the present invention distinguishes the HRM method specificity test of feline enteric coronavirus and feline infectious peritonitis virus
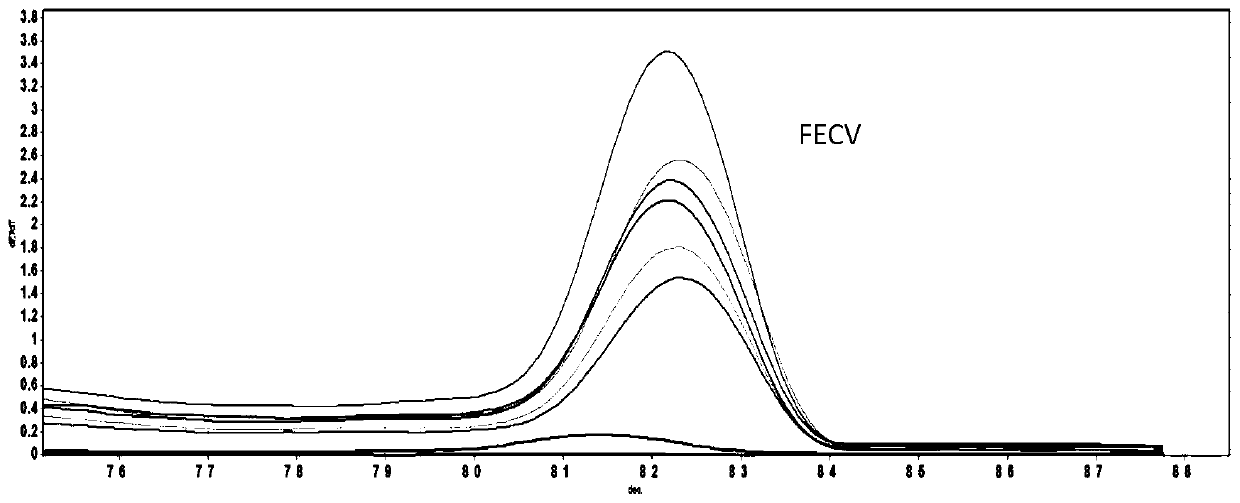
[0050] In order to analyze whether the HRM reaction has detection specificity in other canine and cat pathogens on the basis of being able to distinguish FECV and FIPV, the nucleic acid of the following samples was used as a template for HRM reaction: canine coronavirus (CCV), canine parvovirus (CPV ), canine distemper (CDV), canine adenovirus (CAV), feline herpesvirus (FHV), feline parvovirus (FPV), feline calicivirus (FCV), a negative control (ddH 2 0), one FECV positive sample, one FIPV positive sample.
[0051] The results showed that all samples were negative except for FECV and FIPV (see figure 2 ). It proved that the method has good specificity and no cross-reaction with other pathogens.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com