Genetically-engineered high-producing strain streptomyces diastatochromogenes, production method of epsilon-polylysine and application
A technology of Streptomyces chromogenes and polylysine, applied in the direction of genetic engineering, microorganism-based methods, biochemical equipment and methods, etc., can solve the problem of fewer ε-polylysine-producing bacteria and improve the fermentation level Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
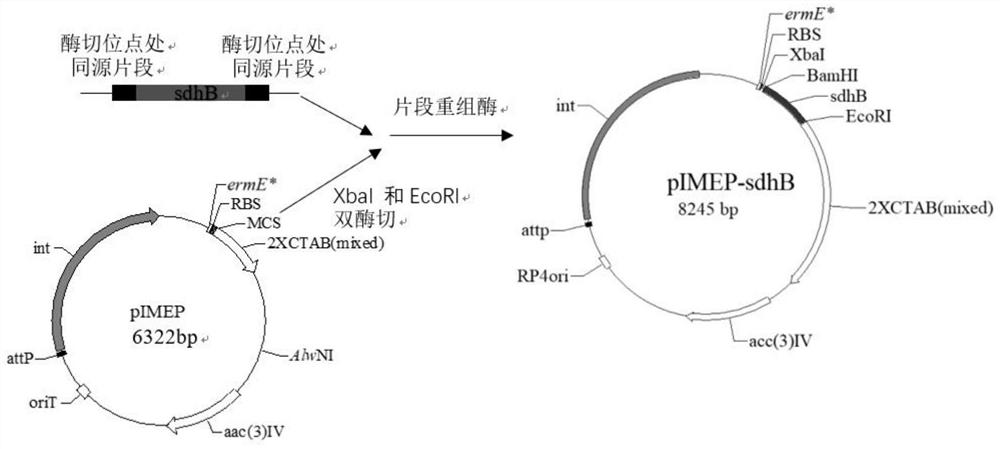
[0077] A kind of genetic engineering high-yield amylase strain Streptomyces chromogenes SDHB (Streptomycesdiastatochromogenes SDHB), the construction steps are as follows:
[0078] (1) Obtaining the target fragment gene:
[0079] The primer sequence sdhB-FF / sdhB-RR was designed according to the sdhB gene, and the restriction sites of XbaI and EcoRI were respectively introduced at both ends of the gene sdhB, and 6 nucleotides were added to the upstream 5' end of the sdhB gene nucleotide sequence, Form the site of the restriction endonuclease XbaI, add 6 nucleotides to the downstream 5' end of the sdhB nucleotide sequence to form the site of the restriction endonuclease EcoRI, PCR amplify the amylase Streptomyces chromogenes TUST sdhB gene in;
[0080] The sequence of the primer sdhB-FF / sdhB-RR is:
[0081] sdhB-FF: SEQ No.2, namely 5'- tctaga atggcgcatgtggaccgg-3', the underlined sequence is the XbaI restriction site;
[0082] sdhB-RR: SEQ No.3, namely 5'- gaattc tcaccgg...
Embodiment 2
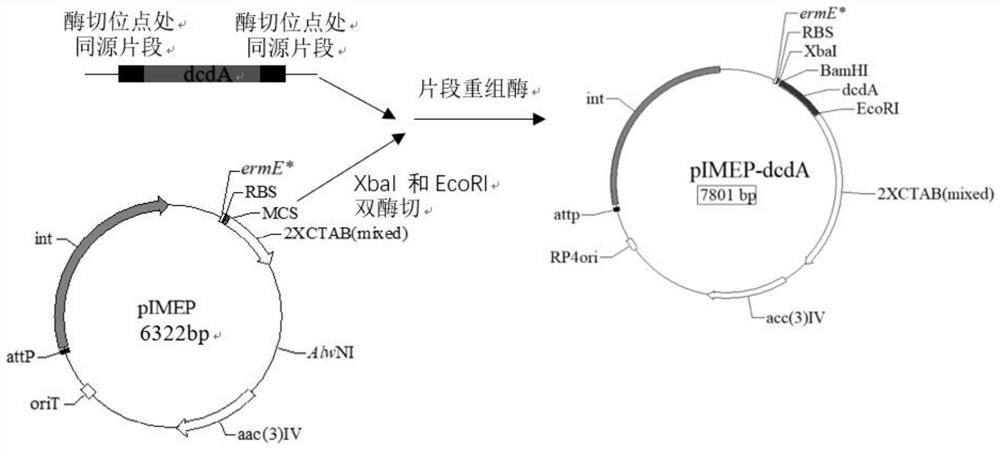
[0094] A kind of genetic engineering high-yielding amylase strain Streptomyces diastatochromogenes DCDA (Streptomycesdiastatochromogenes DCDA), the construction steps are as follows:
[0095] (1) Obtaining the target fragment gene:
[0096] The primer sequence dcdA-FF / dcdA-RR was designed according to the dcdA gene, and the restriction sites of XbaI and EcoRI were respectively introduced at both ends of the gene dcdA, and 6 nucleotides were added to the upstream 5' end of the dcdA gene nucleotide sequence, Form the site of the restriction endonuclease XbaI, add 6 nucleotides to the downstream 5' end of the dcdA nucleotide sequence to form the site of the restriction endonuclease EcoRI, PCR amplify the amylase Streptomyces chromogenes TUST The dcdA gene in;
[0097] The sequence of the primer dcdA-FF / dcdA-RR is:
[0098] dcdA-FF: SEQNo.5, namely 5'- tctaga gtgcctccgtctgtggagc-3', the underlined sequence is the XbaI restriction site;
[0099] dcdA-RR: SEQNo.6, namely 5'- ...
Embodiment 3
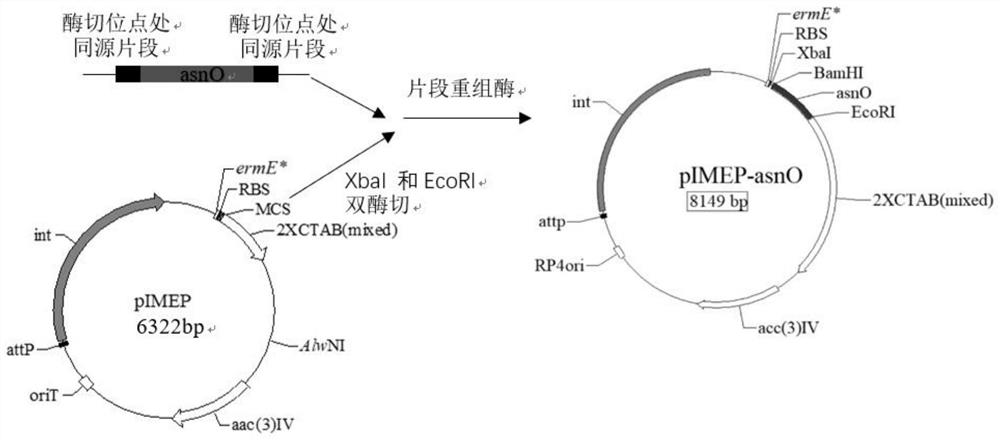
[0111] A kind of genetic engineering high-yield amylase strain Streptomyces chromogenes ASNO (Streptomycesdiastatochromogenes ASNO), the construction steps are as follows:
[0112] (1) Obtaining the target fragment gene:
[0113] The primer sequence asnO-FF / asnO-RR was designed according to the asnO gene, and the restriction sites of XbaI and EcoRI were respectively introduced at both ends of the gene asnO, and 6 nucleotides were added to the upstream 5' end of the asnO gene nucleotide sequence, Form the site of the restriction endonuclease XbaI, add 6 nucleotides to the downstream 5' end of the asnO nucleotide sequence to form the site of the restriction endonuclease EcoRI, PCR amplify the amylase Streptomyces chromogenes TUST asnO gene in;
[0114] The sequence of the primer asnO-FF / asnO-RR is:
[0115] asnO-FF: SEQ No.8, namely 5'- tctaga atgtgcggaatcaccggc-3', the underlined sequence is the XbaI restriction site;
[0116] asnO-RR: SEQ No.9, namely 5'- gaattc tcagagg...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com