Molecular marker for breeding bright-peel tomato varieties and application of molecular marker
A molecular marker and tomato technology, which can be used in applications, microbiological determination/inspection, DNA/RNA fragments, etc., can solve the problems of vitamin and lycopene loss, achieve bright appearance, save production costs, and "good slag removal" Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] The development of molecular markers for the breeding of bright peel or dried and preserved tomato varieties, the specific steps are as follows:
[0034] S1, with glossy as the female parent and tomato wild germplasm LA1375 as the male parent, was crossed to obtain the F1 generation; the F1 generation plants were self-pollinated to obtain the F2 generation.
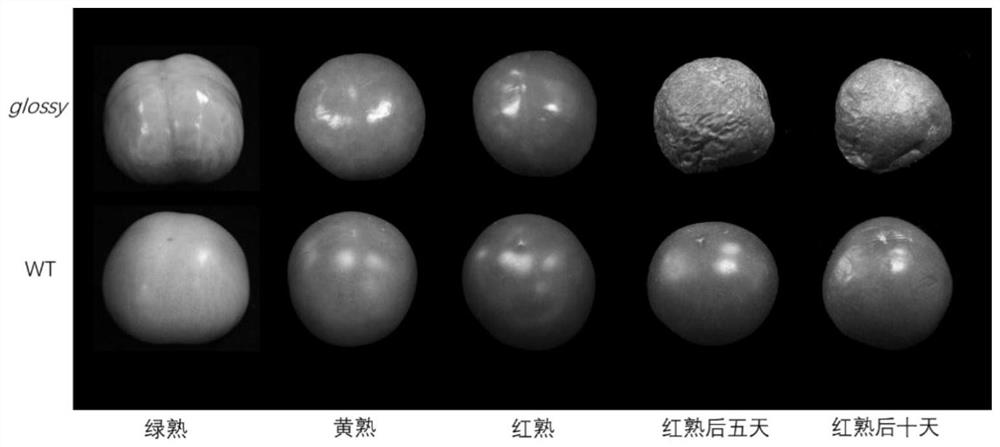
[0035] S2, 563 F2 generation plants were sown and colonized in plastic greenhouses. Under normal management, the phenotype can be basically judged 2-3 weeks after the fruit sets, and the phenotype is obvious and stable during the green ripening period. Investigate more than 3 fruits per plant. The glossy mutant fruit is bright in color and feels sticky when touched, while the normal fruit is matte in color and smooth to the touch. If more than 3 fruits are all bright and sticky, the glossy phenotype is judged to be recessive, otherwise it is dominant.
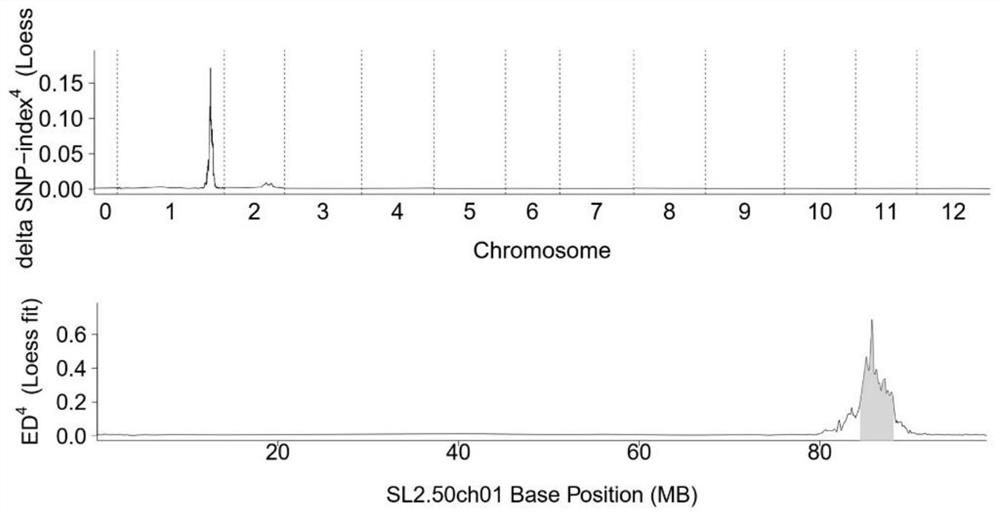
[0036] In the F2 population, there are 138 individual plants...
Embodiment 2
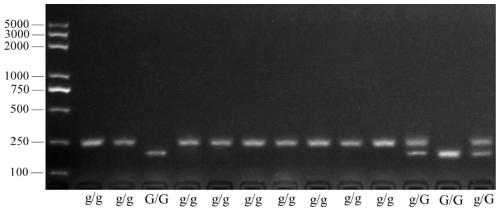
[0056] Using the molecular marker GLS and primer sequences obtained in Example 1, 13 parts of plant materials obtained by glossy mutants as donor materials were identified, and compared with their fruit phenotypes, the specific steps were as follows:
[0057] (1) Using the genomic DNA of related tomato strains as a template, the primers are GLS, and the reaction system and program recommended by Easy Taq polymerase of Beijing Quanshijin Biotechnology Co., Ltd. are used for amplification, and the PCR products are amplified with 2% agarose gel Electrophoretic detection.
[0058] Wherein, the PCR amplification reaction system described in the above technical scheme, according to the total volume of 15 μl, includes the following components:
[0059]
[0060] The PCR amplification reaction conditions were: pre-denaturation at 94.0°C for 3 minutes; denaturation at 94.0°C for 30 s, annealing at 55.0°C for 30 s, extension at 72.0°C for 30 s, and 33 cycles; extension at 72.0°C for 5...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com