Method for detecting novel coronavirus 2019-nCoV by multi-protein joint inspection combination
A coronavirus and combination detection technology, which is applied in the field of multi-protein joint detection combination detection of new coronavirus 2019-nCoV, can solve problems such as false positives and cross-reactions, achieve optimal design, increase sensitivity and specificity, and avoid false negatives and false positives. positive effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] Preparation of test strips:
[0070] 1. Reagent preparation:
[0071] 1.1 Protein dilution: Weigh 55.8g of disodium hydrogen phosphate, 5.5g of sodium dihydrogen phosphate, and 9g of sodium chloride; add 999mL of pure water to dissolve, draw 1mL of proClin300 into the solution, stir and mix, and dilute to 1000mL. Store at room temperature for later use.
[0072] 1.2 Preparation of blocking solution: Weigh 55.8 g of disodium hydrogen phosphate, 5.5 g of sodium dihydrogen phosphate, 9 g of sodium chloride, and 10 g of bovine serum albumin, add 900 mL of pure water to dissolve, add NaOH solution to adjust the pH to 7.4. Draw 1mL proClin300 into the solution, stir and mix, and dilute to 1000mL. Store at 2-8°C for later use.
[0073] 2. Preparation steps:
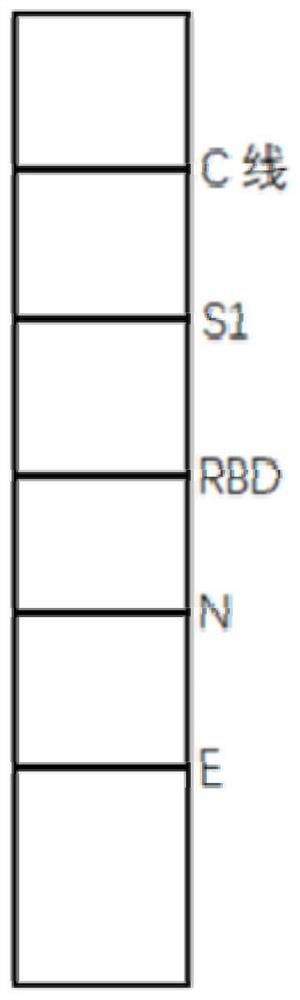
[0074] 2.1 The length and width of the nitrocellulose membrane were cut to 31cm×5cm, and four kinds of high-purity specific recombinant S1 protein, recombinant RBD protein, recombinant N protein, and recombinant E pro...
Embodiment 2
[0090] Preparation of test strips:
[0091] 1. Reagent preparation:
[0092] 1.1 Protein dilution: Weigh 55.8g of disodium hydrogen phosphate, 5.5g of sodium dihydrogen phosphate, and 9g of sodium chloride; add 999mL of pure water to dissolve, draw 1mL of proClin300 into the solution, stir and mix, and dilute to 1000mL. Store at room temperature for later use.
[0093] 1.2 Preparation of blocking solution: Weigh 55.8 g of disodium hydrogen phosphate, 5.5 g of sodium dihydrogen phosphate, and 9 g of sodium chloride; weigh 10 g of bovine serum albumin, add 900 mL of pure water to dissolve, add NaOH solution to adjust the pH to 7.4. Draw 1mL proClin300 into the solution, stir and mix, and dilute to 1000mL. Store at 2-8°C for later use.
[0094] 2. Preparation steps:
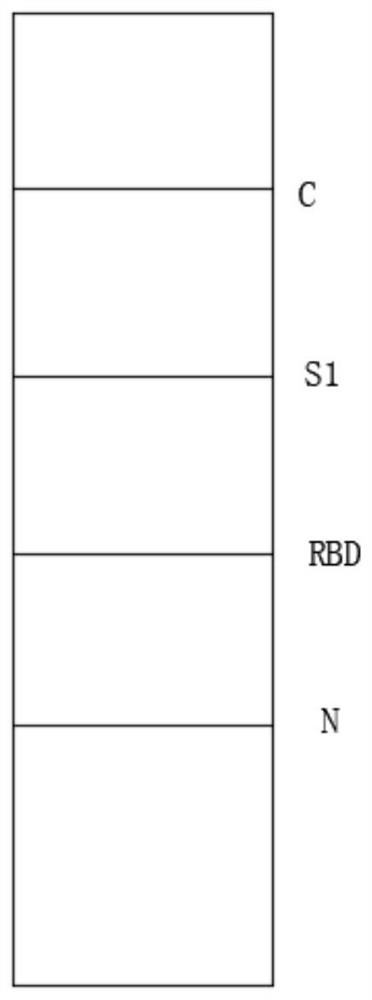
[0095] 2.1 The length and width of the nitrocellulose membrane are cut to 31cm×5cm, and three kinds of high-purity specific recombinant S1 protein, recombinant RBD protein, and recombinant N protein are respecti...
Embodiment 3
[0111] Preparation of test strips:
[0112] 1. Reagent preparation:
[0113] 1.1 Protein dilution: Weigh 55.8g of disodium hydrogen phosphate, 5.5g of sodium dihydrogen phosphate, and 9g of sodium chloride; add 999mL of pure water to dissolve, draw 1mL of proClin300 into the solution, stir and mix, and dilute to 1000mL. Store at room temperature for later use.
[0114] 1.2 Preparation of blocking solution: Weigh 55.8 g of disodium hydrogen phosphate, 5.5 g of sodium dihydrogen phosphate, and 9 g of sodium chloride; weigh 10 g of bovine serum albumin, add 900 mL of pure water to dissolve, add NaOH solution to adjust the pH to 7.4. Draw 1mL proClin300 into the solution, stir and mix, and dilute to 1000mL. Store at 2-8°C for later use.
[0115] 2. Preparation steps:
[0116] 2.1 The length and width of the nitrocellulose membrane are cut to 31cm×5cm, and three kinds of high-purity specific recombinant S1 protein, recombinant RBD protein, and recombinant E protein are respecti...
PUM
| Property | Measurement | Unit |
|---|---|---|
| width | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com