Cardiac troponin I specific nucleic acid aptamer and application thereof
A technology for cardiac troponin and nucleic acid aptamers, which can be used in the biological and medical fields to solve the problems of increased cost, low stability, suboptimal affinity and stability, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Example 1: Synthesis of ssDNA nucleic acid aptamers specifically binding to cTnI protein
[0049] Synthesized by Sangon Bioengineering (Shanghai) Co., Ltd. SEQ ID No.1 (cTnI-14S-7), SEQ ID No.2 (cTnI-14S-3), SEQ ID No.3 (cTnI-14S-16) The indicated nucleic acid aptamers.
Embodiment 2
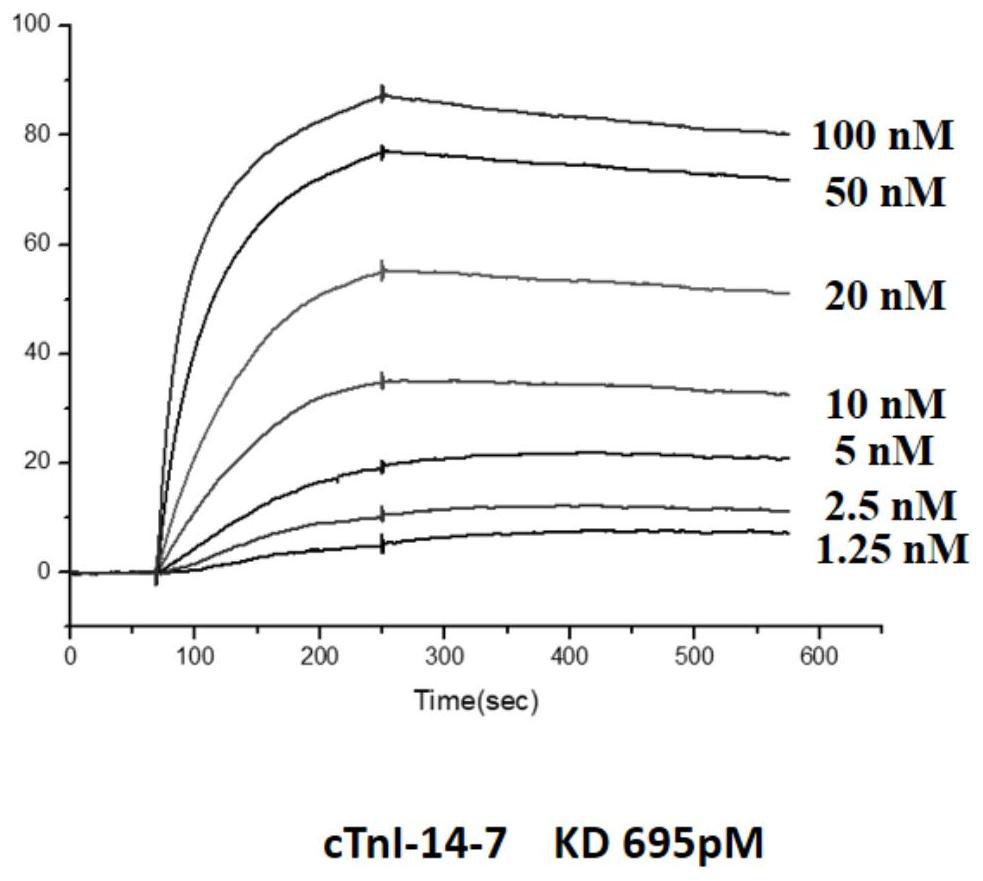
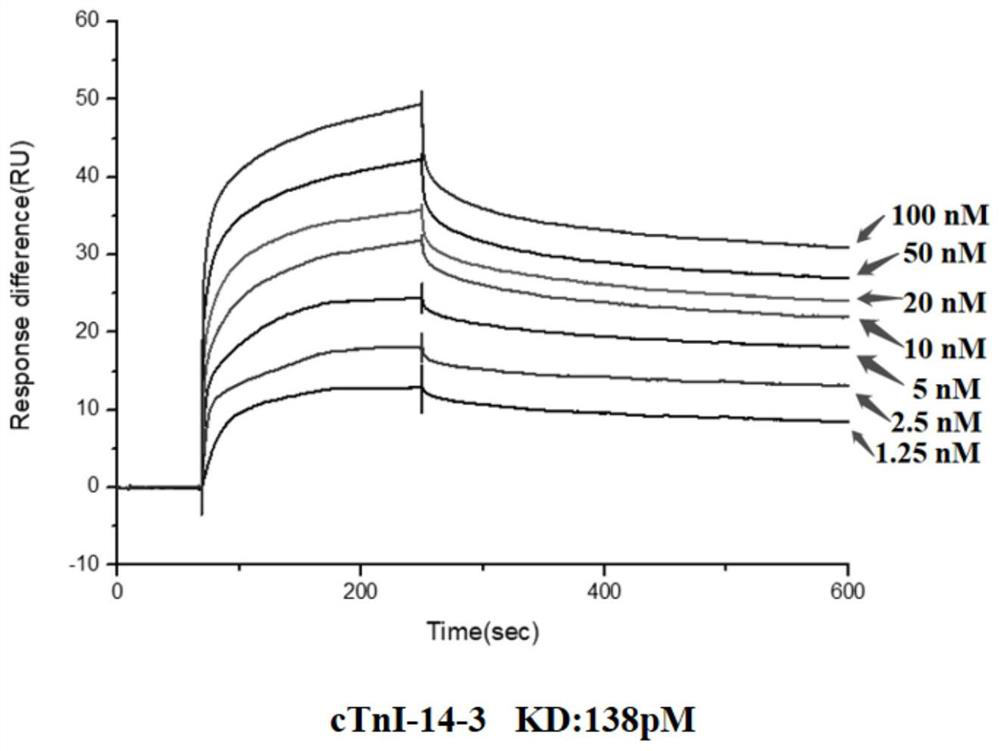
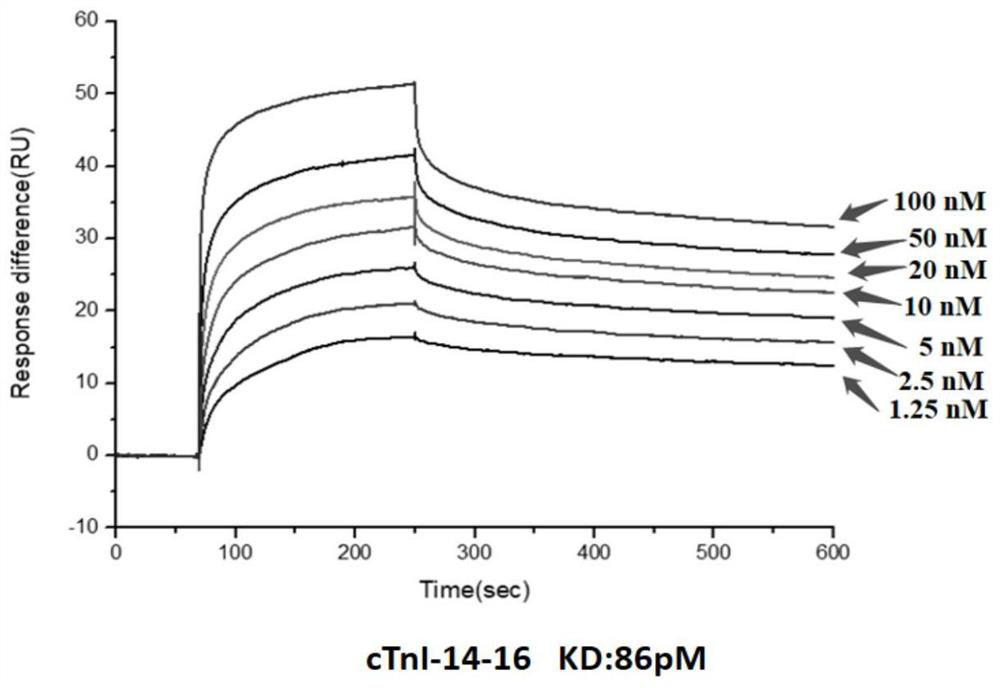
[0050] Embodiment 2: surface plasmon resonance (SPR) detects the affinity of cTnI nucleic acid aptamer and cTnI protein
[0051] 1. The nucleic acid aptamers cTnI-14-7 (SEQ ID No.1), cTnI-14-3 (SEQ ID No.2), cTnI-14-16 (SEQ ID No.3) synthesized by Shanghai Sangon, Diluted with DPBS to: 0, 1.25nM, 2.5nM, 5nM, 10nM, 20nM, 50nM, 100nM;
[0052] 2. Coupling cTnI protein to the surface of CM5 chip (manufacturer: GE Healthcare, model: Biacore Sensor Chipseries CM5): first wash the chip with 50mM NaOH, inject 20ul at a flow rate of 10ul / min, and then use an equal volume of EDC (1 -(3-Dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride; 0.4M aqueous solution) and NHS (N-hydroxysuccinimide; 0.1M aqueous solution) are mixed and injected into 50ul Activate the chip with a flow rate of 5ul / min. The cTnI protein was diluted with 10 mM sodium acetate at pH 4.5 to a final concentration of 50 μg / mL and then injected. The injection volume was 15 μL, the flow rate was 5 uL / min, and the cTn...
Embodiment 3
[0055] Embodiment 3: surface plasmon resonance (SPR) detects the binding situation of cTnI nucleic acid aptamer and interfering protein
[0056] 1. The nucleic acid aptamers cTnI-14-7 (SEQ ID No.1), cTnI-14-3 (SEQ ID No.2), cTnI-14-16 (SEQ ID No.3) synthesized by Shanghai Sangon, Dilute to 100nM with DPBS;
[0057] 2. Coupling cTnI protein, TNFα protein, BSA protein, and SA protein to the 1st, 2nd, 3rd, and 4th channels on the surface of the CM5 chip: first wash the chip with 50mM NaOH, inject 20ul at a flow rate of 10ul / min, and then With equal volumes of EDC (1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride; 0.4M aqueous solution) and NHS (N-hydroxysuccinimide; 0.1M aqueous solution) After mixing the two reagents, inject 50ul of the activated chip at a flow rate of 5ul / min. Dilute the cTnI protein with 10mM sodium acetate at pH 4.5 to a final concentration of 50μg / mL and inject the sample. The injection volume is 15μL, the flow rate is 5uL / min, the cTnI protein ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com