CRISPR-Cas12j enzyme and system
A sequence and protein technology, applied in the field of nucleic acid editing and regularly clustered interspaced short palindromic repeats, can solve problems such as reducing off-target effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0540] Example 1. Acquisition of Cas12j gene and Cas12j guide RNA
[0541] 1. Annotation of CRISPR and genes: Prodigal was used to annotate the microbial genome and metagenomic data of NCBI and JGI databases to obtain all proteins, and Piler-CR was used to annotate CRISPR loci. The parameters were all default parameters.
[0542] 2. Filtering of proteins: De-redundancy of annotated proteins by sequence consistency, removing proteins with completely identical sequences, and classifying proteins longer than 800 amino acids into macromolecular proteins. Since the effector proteins of all the second-type CRISPR / Cas systems found so far are more than 900 amino acids in length, in order to reduce the computational complexity, we only consider macromolecular proteins larger than 800 amino acids when mining CRISPR effector proteins.
[0543] 3. Acquisition of CRISPR-related macromolecular proteins: Extend each CRISPR locus by 10Kb upstream and downstream, and identify the non-redundan...
Embodiment 2
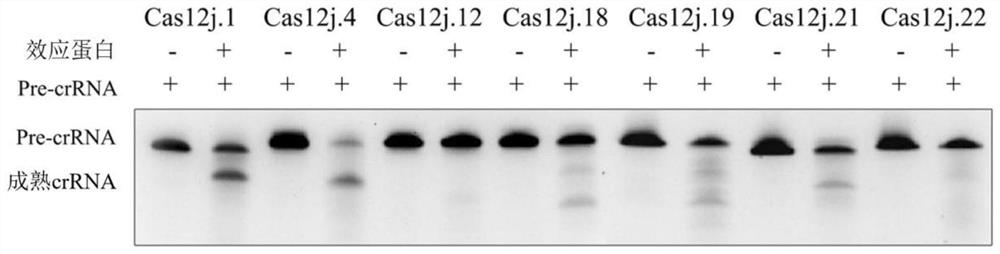
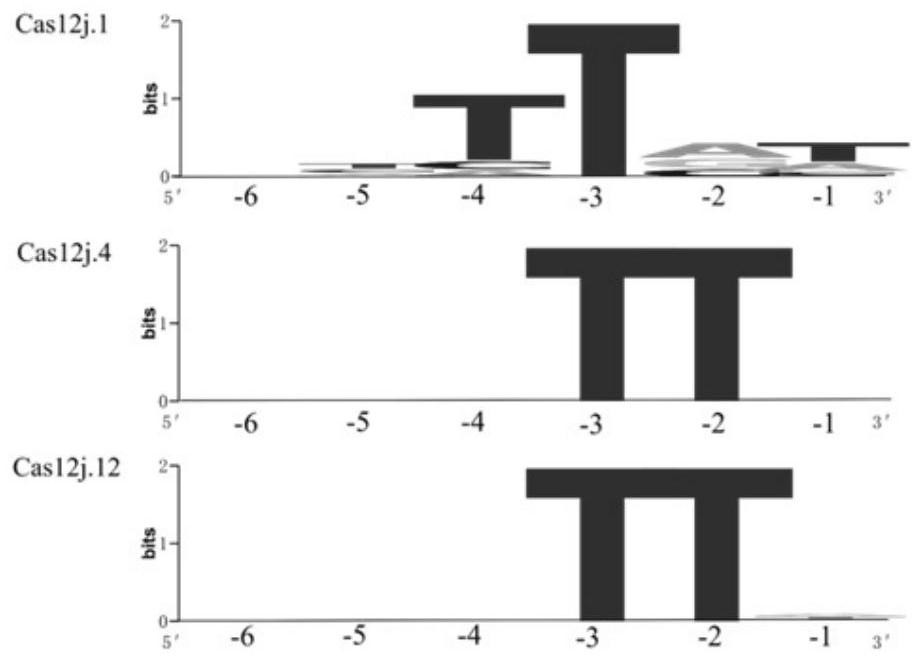
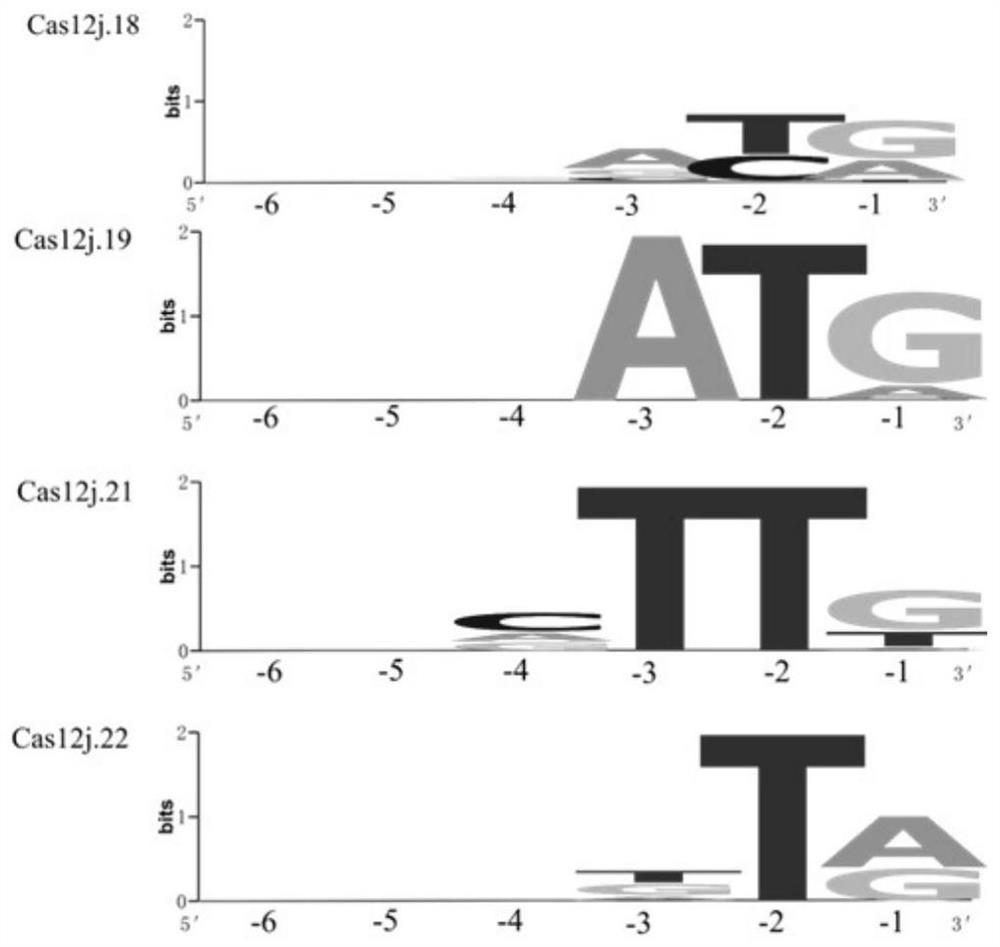
[0548] Example 2. Processing of pre-crRNA by Cas12j gene
[0549] 1. In vitro expression and purification of Cas12j protein
[0550] The steps of in vitro expression and purification of Cas12j protein are as follows:
[0551] 1. Synthesize the DNA sequence encoding Cas12j protein (SEQ ID NO: 82-101) with nuclear localization signal.
[0552] 2. Connect the double-stranded DNA molecule synthesized in step 1 with the prokaryotic expression vector pET-30a(+) to obtain the recombinant plasmid pET-30a-CRISPR / Cas12j.
[0553] 3. The recombinant plasmid pET-30a-CRISPR / Cas12j was introduced into Escherichia coli EC100 to obtain a recombinant strain, which was named EC100-CRISPR / Cas12j.
[0554] A single clone of EC100-CRISPR / Cas12j was taken, inoculated into 100 mL LB liquid medium (containing 50 μg / mL ampicillin), and cultured with shaking at 37 °C and 200 rpm for 12 h to obtain a cultured bacterial liquid.
[0555] 4. Take the cultured bacterial liquid, inoculate it into 50 mL LB...
Embodiment 3
[0609] Example 3. PAM domain identification of Cas12j protein
[0610] 1. The recombinant plasmid pACYC-Duet-1+CRISPR / Cas12j was constructed and sequenced. According to the sequencing results, the structure of the recombinant plasmid pACYC-Duet-1+CRISPR / Cas12j was described as follows: The small fragment between the restriction endonucleases Pml I and Kpn I recognition sequences of the vector pACYC-Duet-1 was replaced with the Cas12j gene ( Double-stranded DNA molecules shown in the sequences shown in SEQ ID NOs: 21-40 from the 1st position from the 5' end to the last position of the 3' end). The recombinant plasmid pACYC-Duet-1+CRISPR / Cas12j expresses the Cas12j protein (SEQ ID NO: 1-20, 107, 108) and the Cas12j guide RNA shown in SEQ ID NO: 104.
[0611] 2. The recombinant plasmid pACYC-Duet-1+CRISPR / Cas12j contains an expression cassette, and the nucleotide sequence of the expression cassette is formed by connecting the Cas12j gene and SEQ ID NO: 104 respectively. For exa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com