Method for carrying out gene mutation on rhodobacter sphaeroides
A technology of Rhodobacter sphaeroides and encoding genes, which is applied in the field of gene manipulation technology and can solve problems such as application limitation, limited number of DNA sequences, and complicated operation process.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0071] Example 1. Using the CRISPR-Cas9 system to mutate a single gene of Rhodobacter sphaeroides
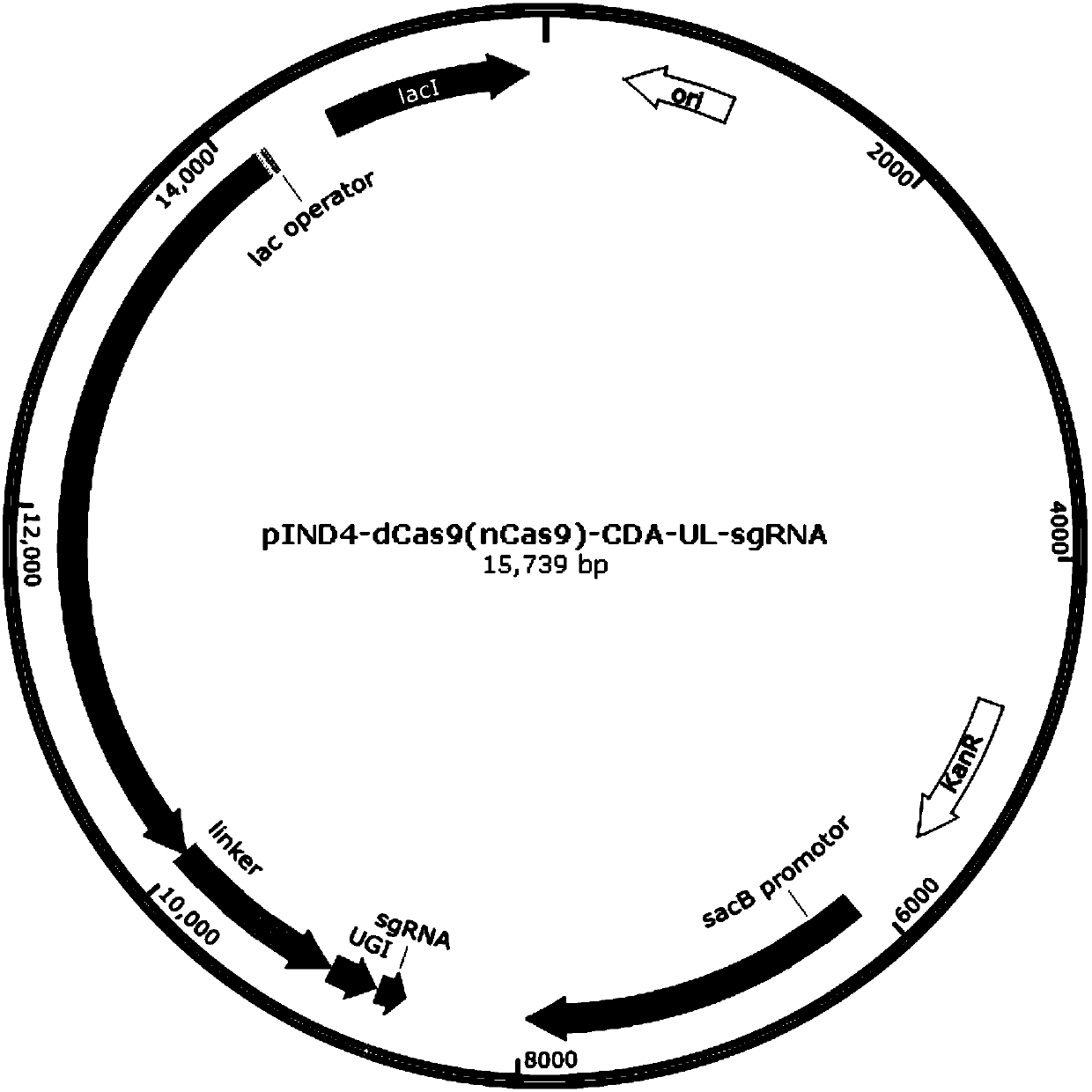
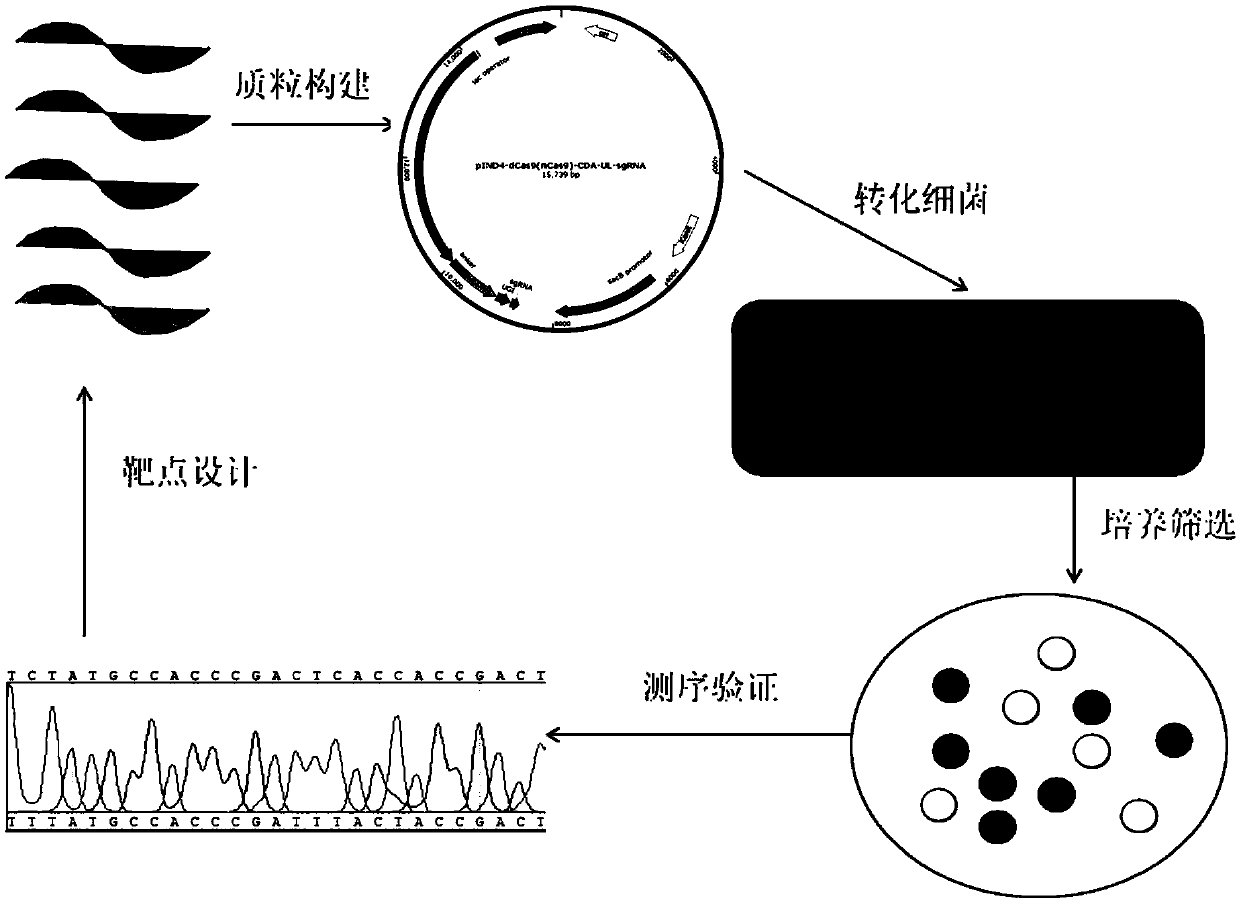
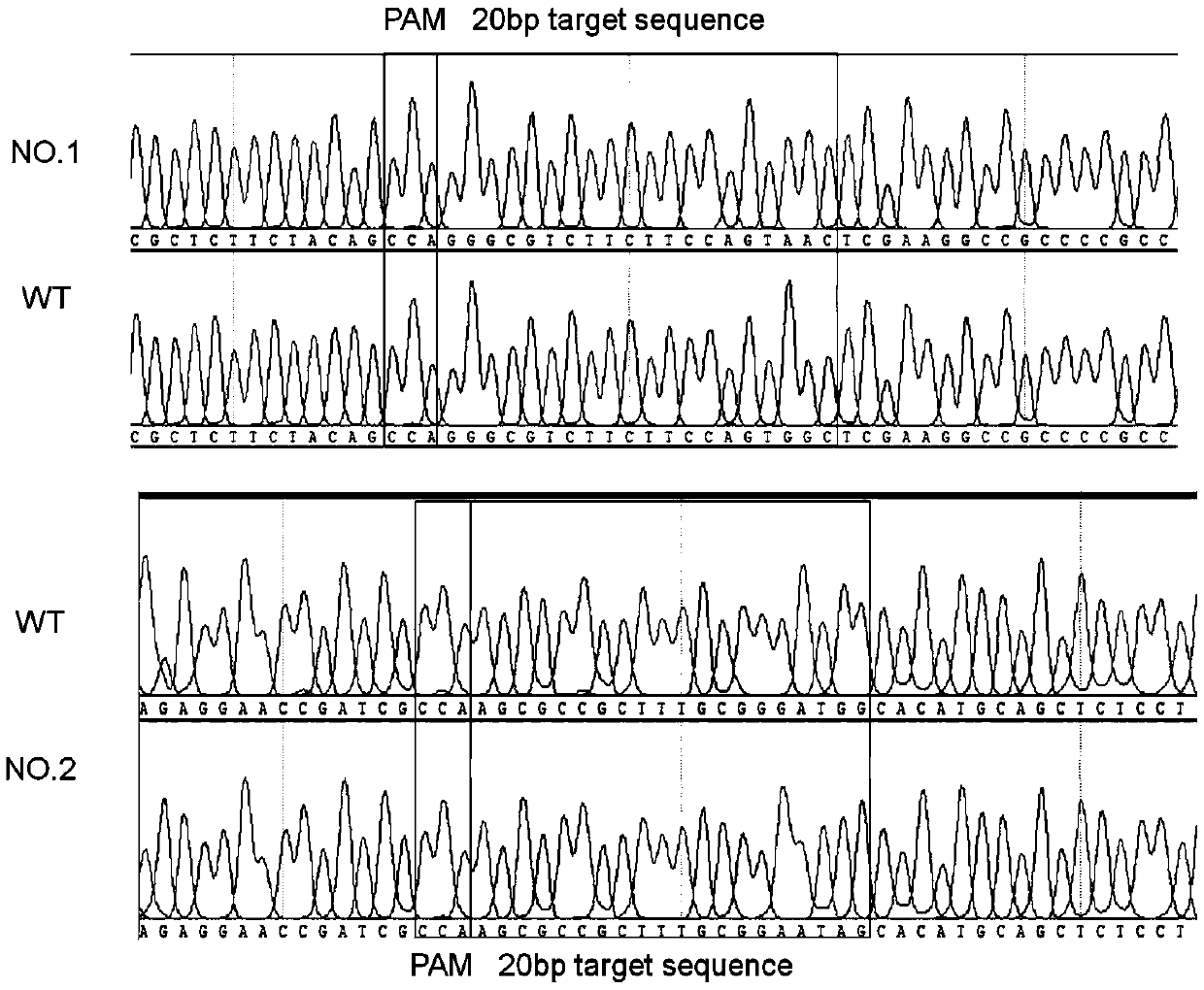
[0072] In this example, the effectiveness of the mutation method described in the present invention was verified by constructing appA, ppsR, and crtB gene mutant strains. Edit the plasmid map as figure 1 As shown, the experimental operation flow chart is as follows figure 2 shown.
[0073] 1. Construction of editing plasmid
[0074] 1. With dCas9 or nCas9 of Streptococcus pyogenes (nCas9 protein is divided into two kinds, i.e. nCas9 (D10A) protein and nCas9 (H840A) protein), the cytosine deaminase pmCDA1 of seven gill mantle (or the adenosine monophosphate of Escherichia coli Deaminase TadA), uracil DNA glycosidase inhibitor UGI of Bacillus subtilis bacteriophage based on the original sequence, codon optimization was carried out on the IDT website (http: / / sg.idtdna.com / codonopt) according to the codon table of Escherichia coli , form new dCas9 or nCas9, pmCDA1 (or TadA), UG...
Embodiment 2
[0113] Example 2. Using the CRISPR-Cas9 system to simultaneously mutate multiple genes of Rhodobacter sphaeroides
[0114] The experimental steps involved in Example 2 are the same as in Example 1, except that the sgRNA in Example 2 is a series of multiple sgRNA expression cassettes, as follows:
[0115] When constructing two or more sgRNAs in tandem, first mix the first two sgRNA1 and sgRNA2 as templates in order, use primers at both ends (sgRNA1-2F / sgRNA2-R) to amplify sgRNA1-sgRNA2, and then use the tandem sgRNA1-sgRNA2 and the following sgRNA3 are mixed as templates, amplified with new double-ended primers (sgRNA1-2F / sgRNA3-R) to obtain sgRNA1-sgRNA2-sgRNA3, and then the tandem sgRNA1-sgRNA2-sgRNA3 and subsequent sgRNA4 are mixed as templates , amplified with new two primers (sgRNA1-2F / sgRNA4-R) to obtain sgRNA1-sgRNA2-sgRNA3-sgRNA4, and so on to obtain several sgRNA series.
[0116] In this embodiment, the specific operation is as follows: use the primer appAsgRNA1-N in ...
Embodiment 3
[0128] Example 3. Using the CRISPR-Cpf1 system to mutate a single gene of Rhodobacter sphaeroides
[0129] This example is the same as Example 1, except that Cas9 is replaced by dCpf1, and sgRNA is replaced by crRNA.
[0130] Wherein the dCpf1 protein is from Francisella noobacillus, the amino acid sequence of the protein is SEQ ID No.2, and the corresponding nucleotide sequence (Escherichia coli codon optimization) is SEQ ID No.8.
[0131] The sequence of the crRNA expression cassette is specifically as follows:
[0132] TTGACAGCTAGCTCAGTCCTAGGTATAATGGATCCGAATTTCTACTGTTGTAGATNNNNNNNNNNNNNNNNNNNNNNNTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTT (SEQ ID No. 14). Wherein, N represents A or T or G or C.
[0133] Wherein, the 1st-35th of SEQ ID No.14 is a promoter; the 36th-55th of SEQ ID No.14 is a repeat sequence; the 56th-79th of SEQ ID No.14 is for expressing spacer DNA sequence; positions 80-119 of SEQ ID No.14 are terminator regions.
[0134] When fused with cytosine deaminase...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com