A method for regulating protein expression based on N-terminal coding sequence modification
A technology for encoding protein and protein expression, applied in biochemical equipment and methods, glycosylase, recombinant DNA technology, etc., can solve problems such as time-consuming, labor-intensive, unsuitable, and unusable
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1: Construction of NCS synonymous mutation library
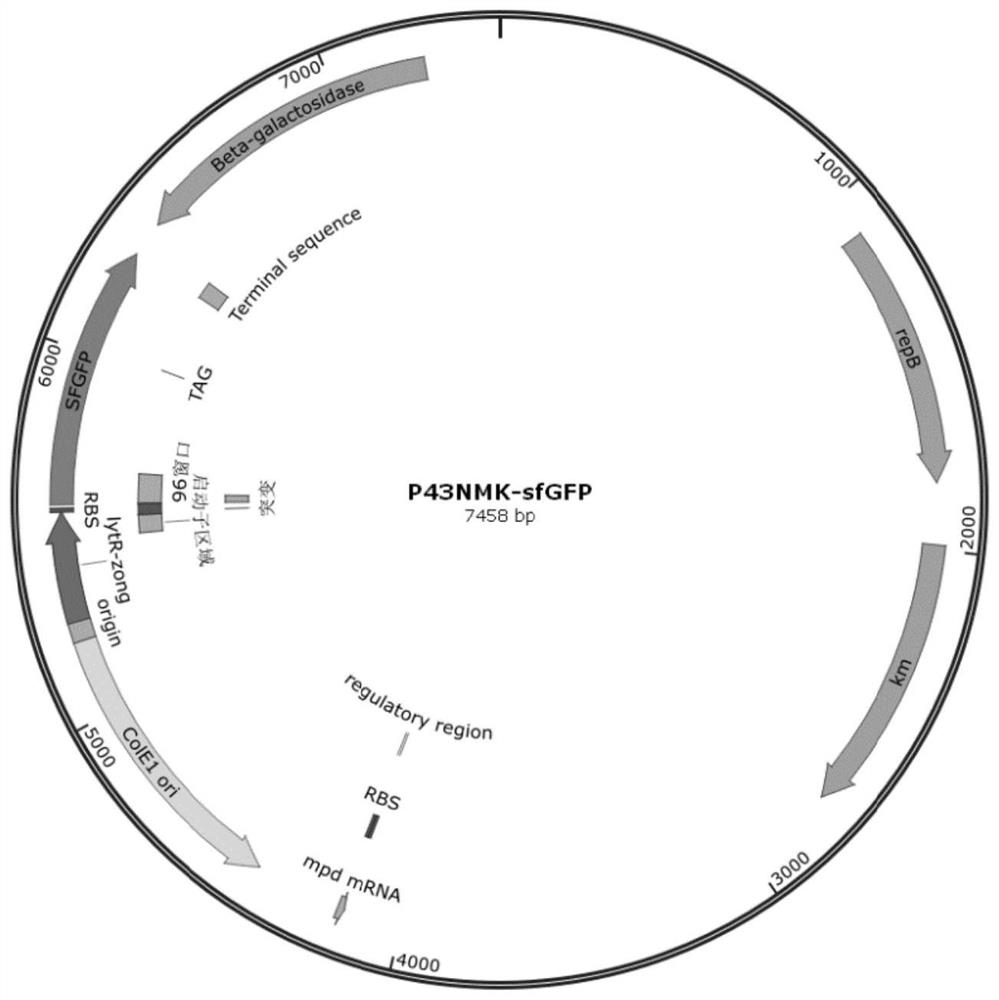
[0050] The PLytr promoter (nucleotide sequence shown in SEQ ID NO.1) was used with primers Lytr-F / Lytr-R (nucleotide sequence shown in SEQ ID NO.2 and 3) and Lytr-F-plasmid / Lytr-R-plasmid (nucleotide sequence shown in SEQ ID NO.4 and 5) was connected to the P43NMK plasmid through a one-step cloning kit, and the plasmid P43NMK-Lytr was constructed;
[0051] Using the same means, the sfGFP fluorescent protein reporter gene (nucleotide sequence shown in SEQ ID NO.6) was used with primers sfGFP-F / sfGFP-R (nucleotide sequence shown in SEQ ID NO.7 and 8) and sfGFP-F-plasmid / sfGFP-R-plasmid (nucleotide sequence shown in SEQ ID NO.9 and 10), through a one-step cloning kit fused to the downstream of PLytr, to obtain the construction of P43NMK-Lytr_sfGFP, such as figure 1 shown;
[0052] Using P43NMK-Lytr_sfGFP as a template, using the degenerate primers sfGFP-F-NCS / sfGFP-R-NCS (nucleotide sequence shown in SEQ ID NO...
Embodiment 2
[0053] Example 2: Characterization of the NCS synonymous mutation library
[0054] The recombinant plasmids with synonymous mutations constructed in Example 1 were respectively transformed into the expression host Bacillus subtilis WB600, and the transformed single clones were inoculated into 96 shallow-well plates containing 200 μL of LB seed medium, and cultured for 8 hours;
[0055] Next, inoculate into a 96-deep-well plate containing 800 μL TB medium according to the inoculum amount of 4 mL / 100 mL, and cultivate for 24 hours to obtain a fermentation broth;
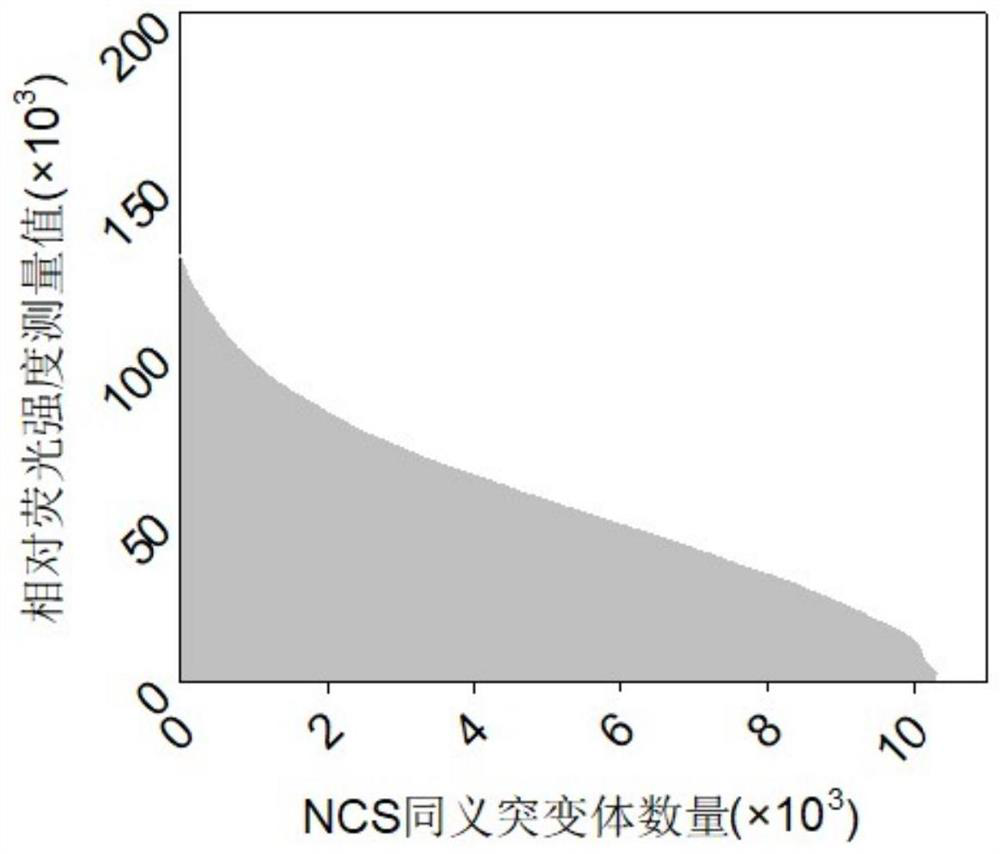
[0056] Then the fermented liquid was quickly placed on ice to freeze, after centrifugation, the supernatant was removed, diluted to an appropriate multiple with PBS buffer (100mM, pH7.2), and passed through a Cytation3 cell imaging microplate detector (U.S. Boten Instrument Co., Ltd. company) to measure the fluorescence value (excitation light 480, absorption light 520) and OD 600 . A total of 8598 single colonies we...
Embodiment 3
[0057] Example 3: Sequence Identification and Fermentation of Representative Samples
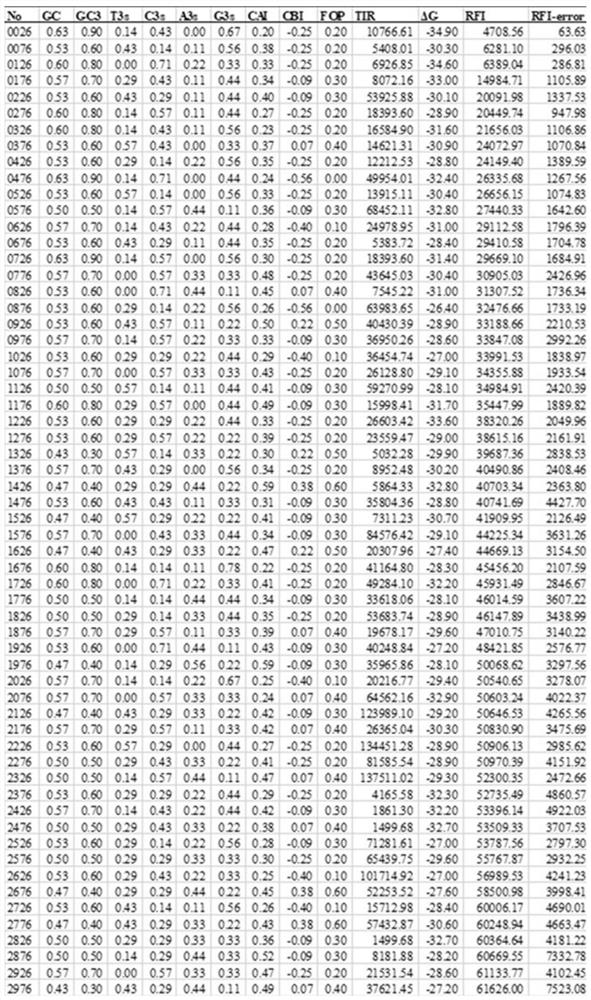
[0058] In Example 2, a total of 8,598 monoclonal host cells were characterized, and the fluorescence value / OD was defined as the relative fluorescence intensity RFI. According to the level of the RFI value, the monoclonal cells were sorted from high to low, and one sequence identification was selected for every 50 (that is, the first One of the 1st to 50th strains was selected, one of the 51st to 100th strains was selected, and so on), and a total of 172 single clones were identified by sequencing.
[0059] Inoculate 172 single clones identified by sequencing into a 250mL shake flask containing 20mL seed medium, and ferment at 37°C and 220rpm for 8 hours to OD 600 If it is greater than 4, inoculate it into a 250mL shake flask containing 25mL fermentation medium according to the ratio of 4mL / 100mL, and measure the fluorescence value and OD of sfGFP after 24 hours of fermentation 600 . Three...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com