Hi-C high-throughput sequencing library building method suitable for microbial metagenomics and application
A metagenomics and metagenomics technology, which is applied in the field of Hi-C high-throughput sequencing library construction of microbial metagenomics, can solve the problems of single cell library construction schemes, etc., to improve the effective data rate and expand the scope of application Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
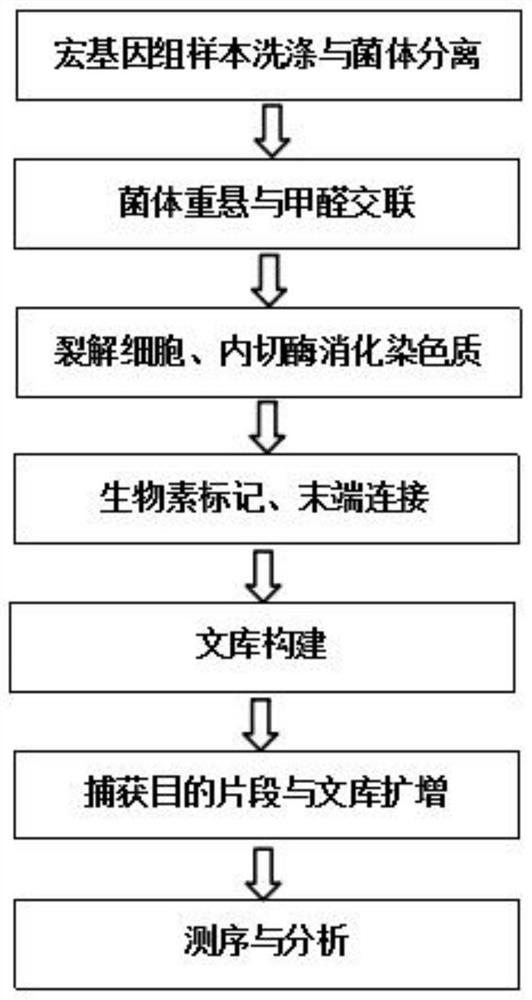
[0051] In this example, soil microorganisms were used as the research object, and metagenomics Hi-C high-throughput sequencing was carried out to build and analyze the library. The specific experimental process is as follows:
[0052] 1. Formaldehyde fixation
[0053] 1) Take 1g of fresh soil / intestinal microorganism / marine or river sediment and other metagenomic samples, add 10mL of 1×PBS and mix well;
[0054] 2) Add 1mL of LB medium and mix well, so that the bacteria have sufficient nutrition to ensure immortality;
[0055] 3) Place the centrifuge tube vertically, let it settle naturally for 30 minutes, let the large sand and gravel settle in the lower layer, and the bacteria will swim in the upper layer solution when they are active, and absorb the upper layer solution into a new centrifuge tube;
[0056] 4) Centrifuge at 500g RT for 5min, draw the upper layer solution into a new centrifuge tube;
[0057] 5) Centrifuge at 12000g RT for 5min, and discard the supernatant; ...
Embodiment 2
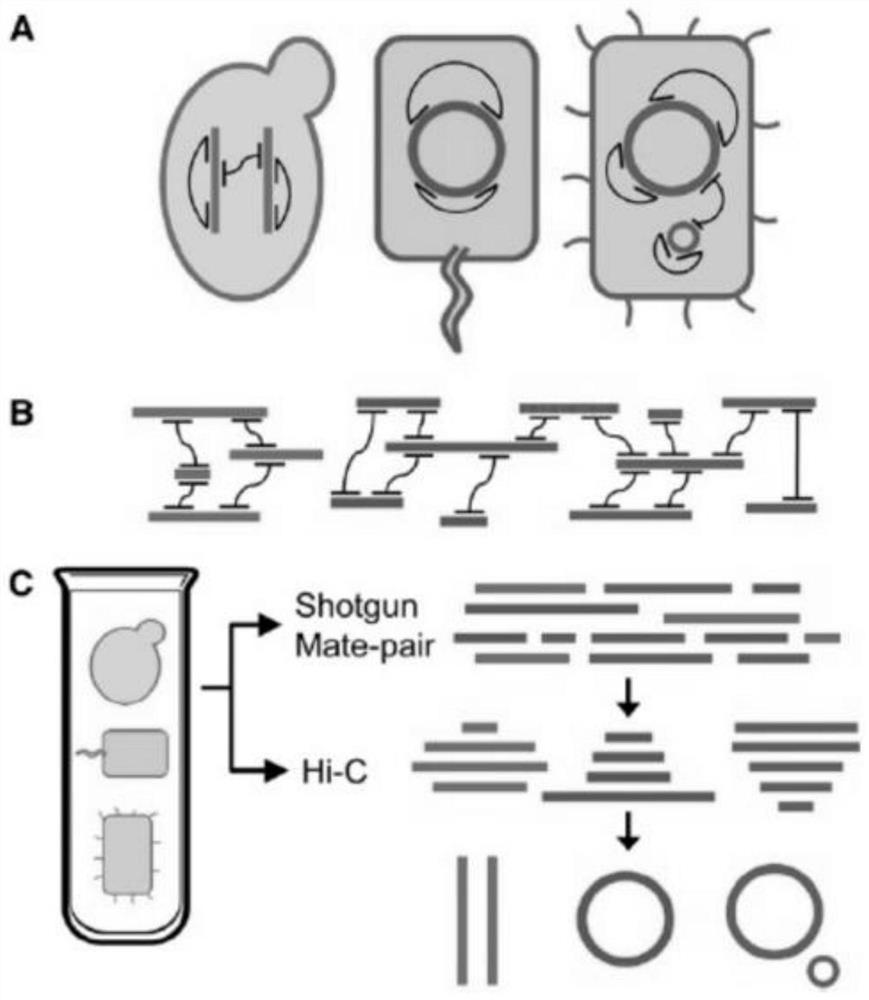
[0172]In this example, animal excrement was used as the research object of intestinal microorganisms, and metagenomics Hi-C high-throughput sequencing was carried out to build and analyze the intestinal microbial samples. Cells release chromatin, and then undergo steps such as digestion of chromatin, biotin labeling, end connection, library construction, capture of target fragments, and amplification to achieve Hi-C high-throughput sequencing library construction for intestinal microbial metagenomics.
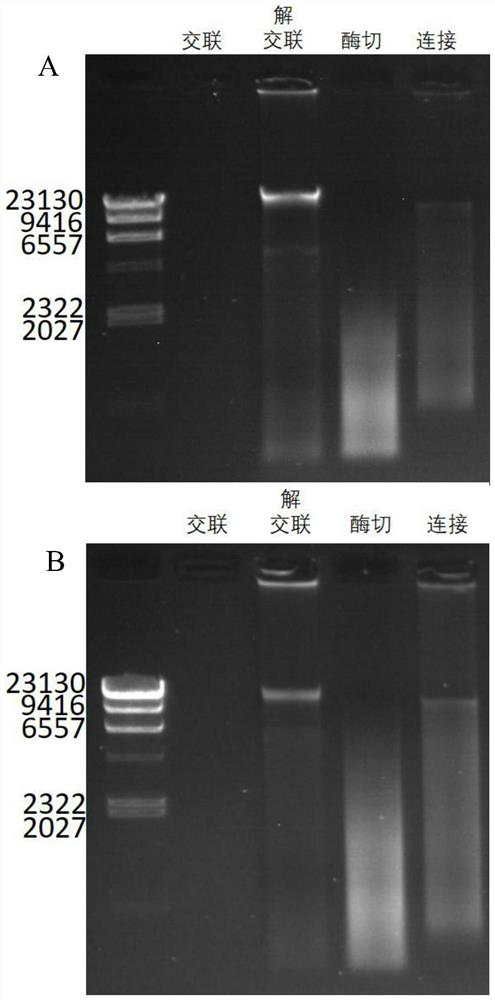
[0173] Among them, the genome integrity, enzyme digestion effect and connection effect are identified by agarose gel electrophoresis pattern, and the agarose gel electrophoresis pattern is as follows: image 3 As shown in B, the decrosslinked electrophoresis strip of the metagenomic group in the figure shows degradation and tailing. Since the metagenome contains bacteria of various physiological stages, it is normal for the decrosslinked DNA to be slightly degraded, and the enti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com