Group of SSR molecular markers based on camellia japonica transcriptome and application of SSR molecular markers in camellia plants
A technology of molecular markers and transcriptomes, applied in the biological field, can solve the problems that the Camellia plants cannot be widely used, the application effect of the Camellia plants cannot be expected, and the average PIC value is low, so as to achieve high polymorphism, high specificity, high metastatic effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
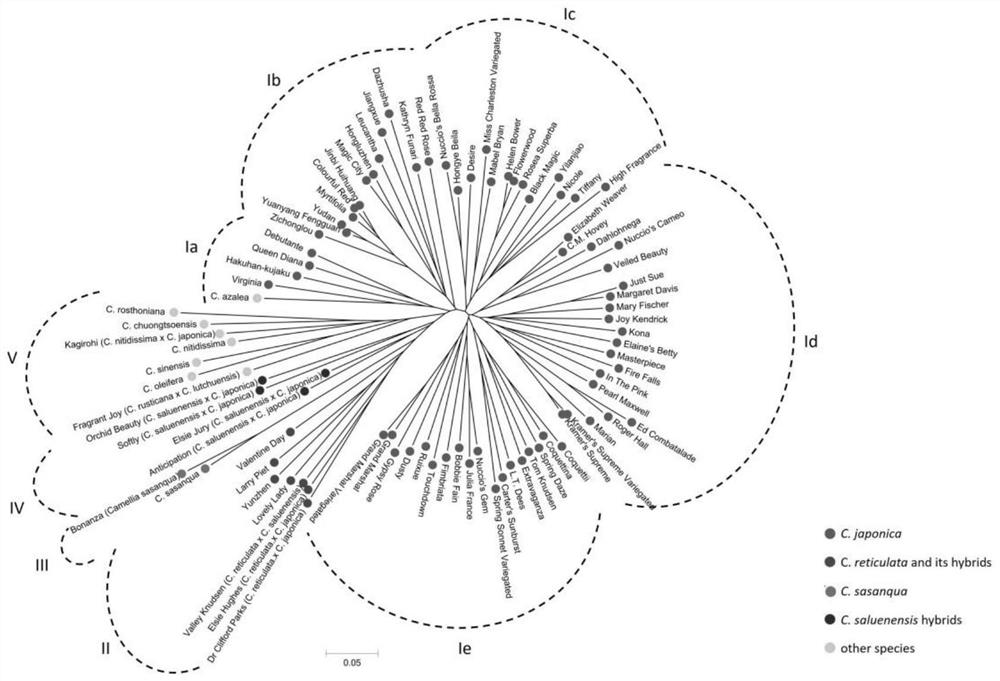
[0035] The inventors of the present application previously obtained high-quality leaf transcriptomes of Camellia japonica (C. japonica) through RNA sequencing technology based on the Illumina platform. These sequence data provide a good resource for the development of Camellia SSR markers. The purpose of this study is to: 1) develop a set of SSR molecular markers based on the Camellia transcriptome; 2) determine the generality and polymorphism of these SSR markers in Camellia species; 3) reveal the genetic relationship and genetic structure of Camellia . The results of this study will provide important information for genetic improvement studies such as taxonomic studies, genetic diversity analysis, molecular marker-assisted breeding, and SSR-based genetic linkage map construction of Camellia plants.
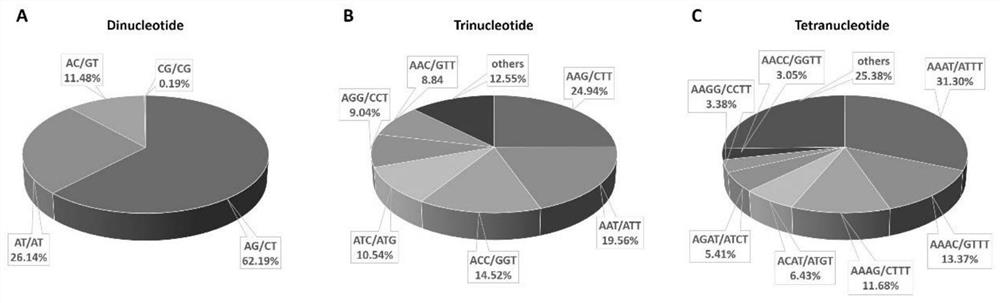
[0036] 1. Transcriptome SSR characteristics of Camellia japonica leaves
[0037] SSRs are highly abundant in the assembled Camellia japonica leaf transcriptome. This applicat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com