Marker for predicting prognosis of colon cancer and application of marker
A colon cancer, marker technology, applied in the field of genetic technology and medicine, can solve the problem of ignoring clinical pathology and risk factors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1 Screening of autophagy-related genes for prognosis prediction of colorectal cancer
[0046] Gene expression datasets and clinical information of COAD patient samples (41 normal samples and 473 tumor samples) were used from the publicly available TCGA dataset. The validation set used 124 colon cancer samples from the GEO dataset GSE72970. Autophagy genes were screened from the human autophagy database HADb to obtain 231 differentially expressed autophagy genes. Univariate Cox regression analysis was used in R software to evaluate whether autophagy-related RNAs were associated with OS. mRNAs with |log2FC|>1 and p2.5 and p1 were defined as risk genes.
[0047] Screening results: Differentially expressed autophagy-related mRNA heatmaps are shown in the accompanying drawings figure 1 As shown in A, the forest plot of the prognosis of autophagy-related mRNA is shown in figure 1 Shown in B; the heat map of differentially expressed autophagy-related lncRNAs is s...
Embodiment 2
[0048] Example 2 Establishing an autophagy-related prognostic risk model and using the risk model to predict the prognostic risk of colon cancer patients
[0049] Risk models were constructed separately for prognostic-related autophagy mRNAs and lncRNAs using multivariate Cox regression. The model analyzes each patient's risk score by incorporating the expression values for each specific RNA, weighted by its estimated regression coefficient. According to the risk score formula, patients were divided into low-risk group and high-risk group with the median risk as the cut-off point. Survival differences between the two groups were assessed by KM and compared using the log-rank statistical method. Multivariate Cox regression analysis and stratified analysis were used to examine the role of the risk score in predicting patient outcomes.
[0050] The autophagy-related mRNA risk model (model 1) was constructed using the TCGA colon cancer dataset (colon cancer; n=514: 41normal sa...
Embodiment 3
[0066] Embodiment 3 establishes a nomograph based on a risk model (obtained by constructing a COX model)
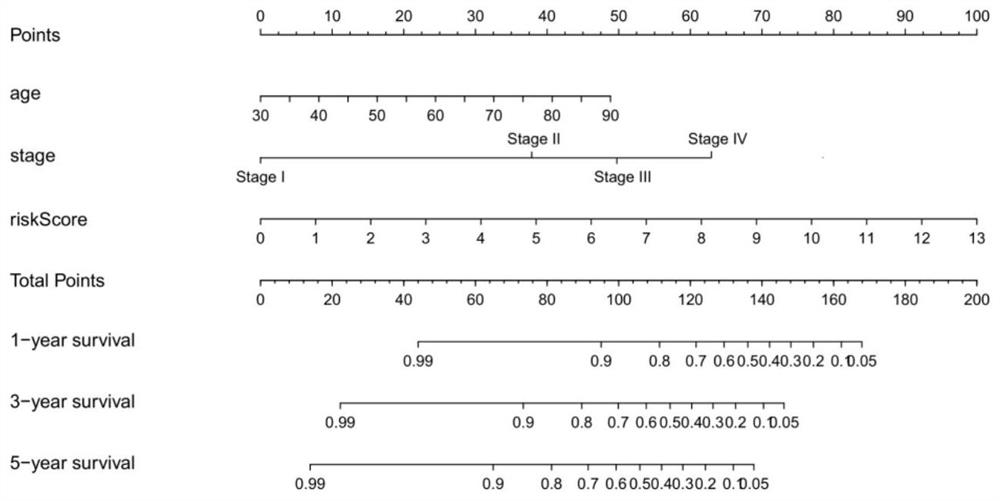
[0067] Construction of a risk scoring system incorporating mRNA, NOMO of age and tumor stage figure 1 ;
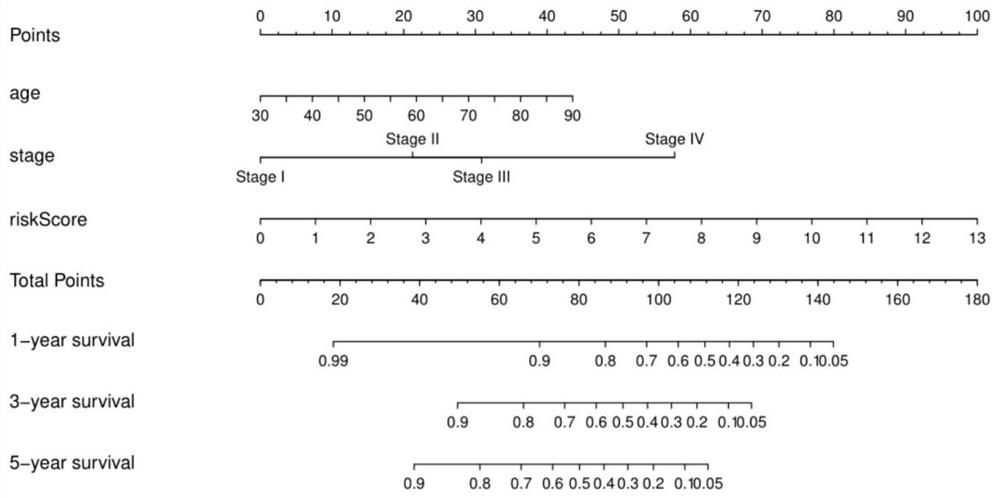
[0068] Construction of a risk scoring system combining 14 lncRNAs, nomo of age and tumor stage figure 2 ;
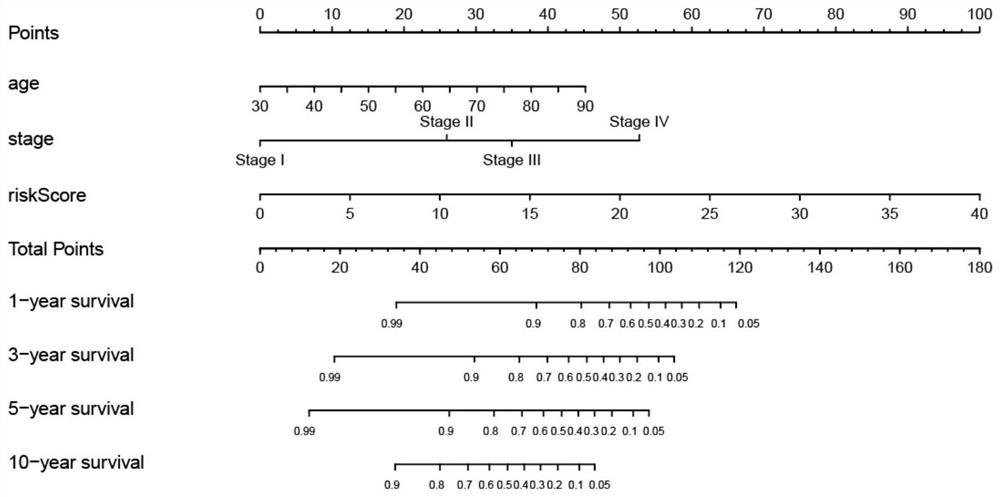
[0069] Construct a risk scoring system combining 6 mRNAs and 14 lncRNAs and a nomo of age and tumor stage image 3 ;
[0070] According to the clinical test results, the scores are listed (Nomo Figure 1-3 The "risk score" in is calculated according to the formulas of the above models 1-3) to predict the 1-year, 3-year, 5-year or 10-year survival rate of each patient.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com