Preparation and application of homologous recombination repair detection reference substance
A technology of homologous recombination and detection reference, which is applied in the field of preparation of cancer cell detection reference products, can solve the problems of self-evident importance and influence on the approval process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054]A homologous recombination repair detection reference product, the cell line used is the tumor cell line KBM-7, including 25 kinds of homologous recombination repair related genes with inactivating mutations, namely: ATM, ATR, BARD1, BLM, BRCA1 , BRCA2, BRIP1, CDK12, CHEK2, FANCA, FANCC, FANCD2, FANCE, FANCF, FANCI, FANCL, FANCM, NBN, PALB2, PPP2R2A, RAD51B, RAD51D, RAD50, RPA1, and RAD54L.
Embodiment 2
[0055] Example 2: A method for preparing a homologous recombination repair detection reference product (germline mutation)
[0056] Step S1: Perform gene editing on the selected tumor cell line. The target genes for gene editing include 25 genes related to homologous recombination repair, namely: ATM, ATR, BARD1, BLM, BRCA1, BRCA2, BRIP1, CDK12, CHEK2, FANCA , FANCC, FANCD2, FANCE, FANCF, FANCI, FANCL, FANCM, NBN, PALB2, PPP2R2A, RAD51B, RAD51D, RAD50, RPA1 and RAD54L, the detailed operation is as follows:
[0057] Step S11, design a specific gRNA for the target site of the target gene, construct a gRNA expression vector, design and synthesize single-stranded DNA, and use it as a template for gene editing and repair;
[0058] Step S12, co-transfecting the gRNA expression vector and the single-stranded DNA into the target cells;
[0059] Step S13, when the confluence of the cells in step S12 reaches 70%-90%, digest and collect the cells, count the cells accurately, and dilute ...
Embodiment 3
[0095] Example 3: A method for preparing a homologous recombination repair detection reference product (somatic mutation)
[0096] In this example, the tumor cell line KBM-7 is used for gene editing, gene and mutation site list, gDNA extraction method, probe list and ddPCR method as described in Example 2, because the frequency of somatic mutations is generally low , so the preparation of this reference product (single tube) was carried out by means of gDNA mixing.
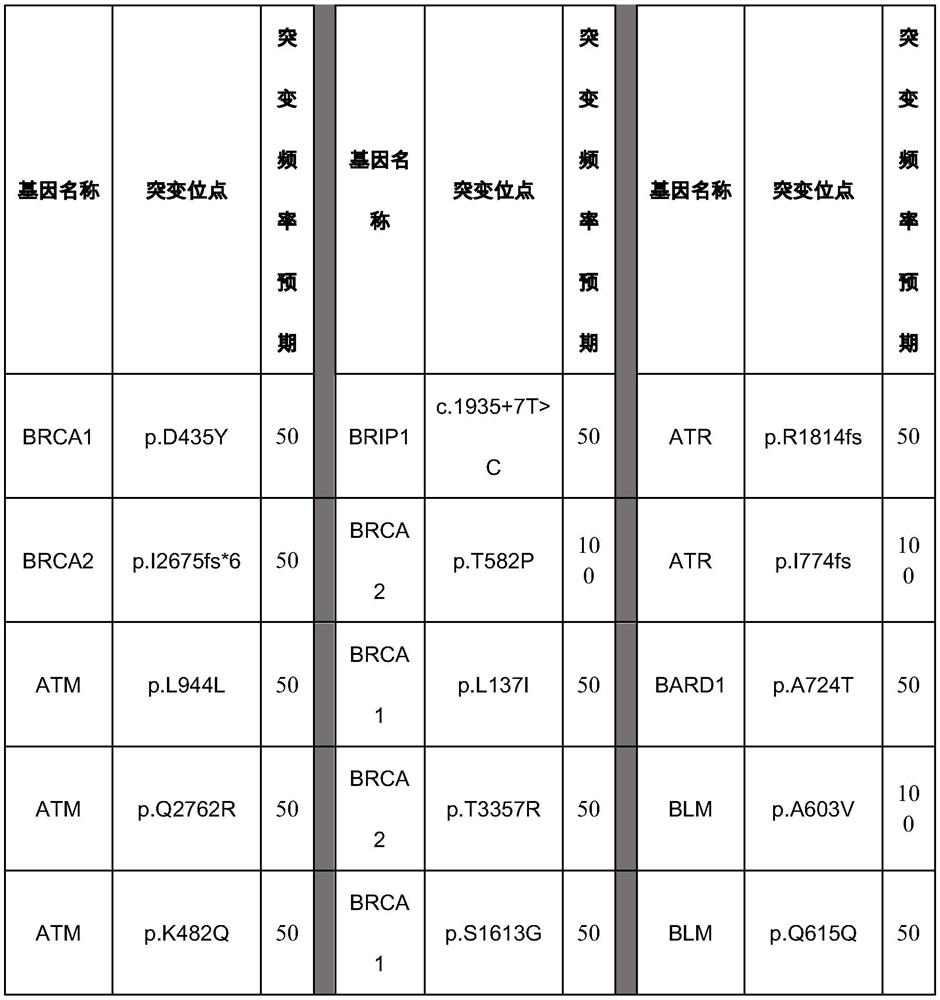
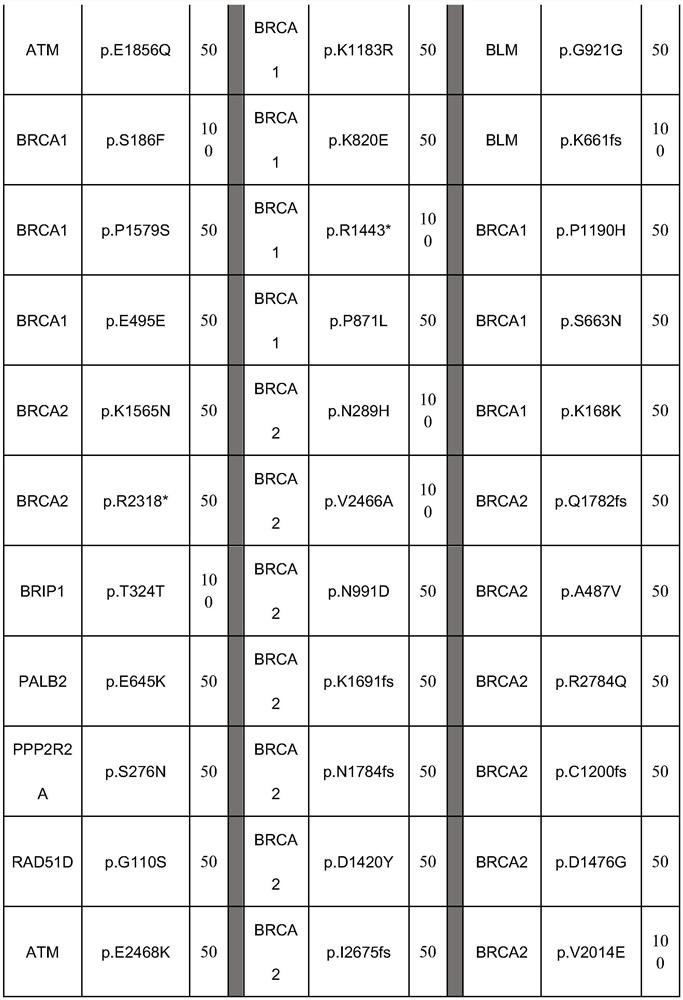
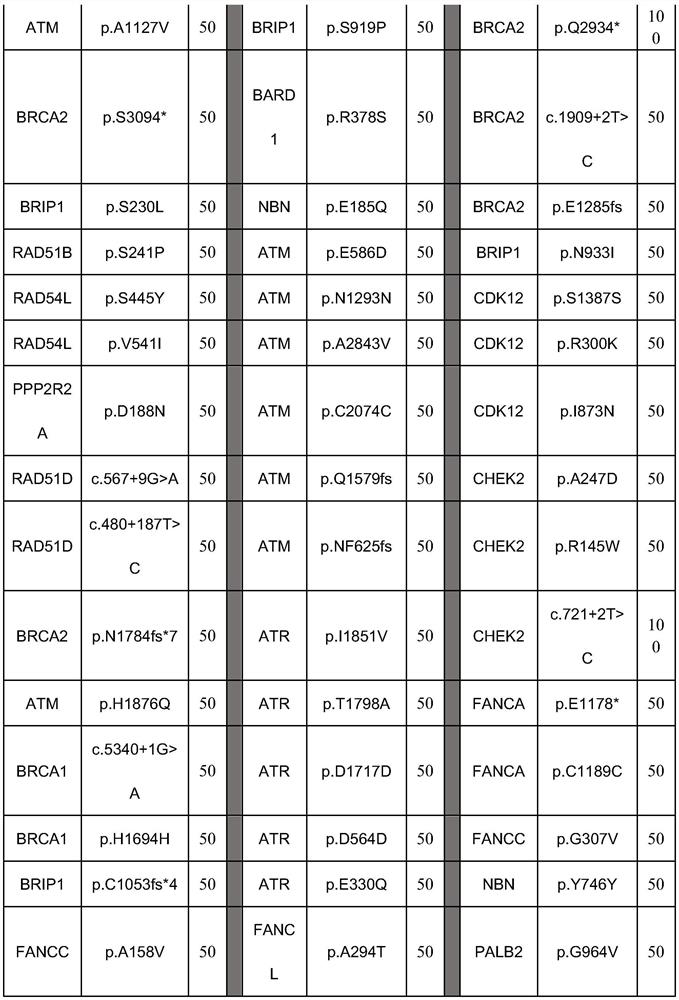
[0097] Select 27 components of somatic mutation reference products in combination of all mutation sites, and the detailed site information has been verified by ddPCR as shown in the table below:
[0098]
[0099]
[0100] This combination forms a somatic mutation reference for genes involved in homologous recombination repair.
[0101] The homologous recombination repair detection reference product prepared above can be used in the detection of homologous recombination repair defects.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com